2-hydroxy-dATP diphosphatase

| 2-hydroxy-dATP diphosphatase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.6.1.56 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
2-hydroxy-dATP diphosphatase (EC 3.6.1.56, also known as oxidized purine nucleoside triphosphatase, or (2'-deoxy) ribonucleoside 5'-triphosphate pyrophosphohydrolase, or Nudix hydrolase 1 (NUDT1), or MutT homolog 1 (MTH1), or 7,8-dihydro-8-oxoguanine triphosphatase) is an enzyme that in humans is encoded by the NUDT1 gene.[1][2][3] During DNA repair, the enzyme hydrolyses oxidized purines and prevents their addition onto the DNA chain. As such it has important role in aging and cancer development.
Function
[edit]This enzyme catalyses the following chemical reaction
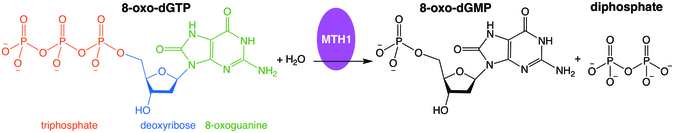
- 2-hydroxy-dATP + H2O 2-hydroxy-dAMP + diphosphate
The enzyme hydrolyses oxidized purine nucleoside triphosphates. The enzyme is used in DNA repair, where it hydrolysis the oxidized purines and prevents their addition onto the DNA chain.[4]
Misincorporation of oxidized nucleoside triphosphates into DNA and/or RNA during replication and transcription can cause mutations that may result in carcinogenesis or neurodegeneration. First isolated from Escherichia coli because of its ability to prevent occurrence of 8-oxoguanine in DNA,[5] the protein encoded by this gene is an enzyme that hydrolyzes oxidized purine nucleoside triphosphates, such as 8-oxo-dGTP, 8-oxo-dATP, 2-oxo-dATP, 2-hydroxy-dATP, and 2-hydroxy rATP, to monophosphates, thereby preventing misincorporation.
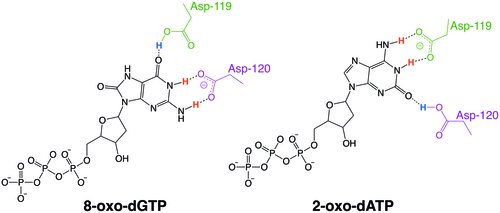
MutT enzymes in non-human organisms often have substrate specificity for certain types of oxidized nucleotides, such as that of E. coli, which is specific to 8-oxoguanine nucleotides. Human MTH1, however, has substrate specificity for a much broader range of oxidatively damaged nucleotides. The mechanism of hMTH1's broad specificity for these oxidized nucleotides is derived from their recognition in the enzyme's substrate binding pocket due to an exchange of protonation state between two nearby aspartate residues.[6]
The encoded protein is localized mainly in the cytoplasm, with some in the mitochondria, suggesting that it is involved in the sanitization of nucleotide pools both for nuclear and mitochondrial genomes. In plants, MTH1 has also been shown to enhance resistance to heat- and paraquat-induced oxidative stress, resulting in fewer dead cells and less accumulation of hydrogen peroxide.[7]
Several alternatively spliced transcript variants, some of which encode distinct isoforms, have been identified. Additional variants have been observed, but their full-length natures have not been determined. A single-nucleotide polymorphism that results in the production of an additional, longer isoform has been described.[3]
Research
[edit]Aging
[edit]A mouse model has been studied that over-expresses hMTH1-Tg (NUDT1).[8] The hMTH1-Tg mice express high levels of the hMTH1 hydrolase that degrades 8-oxodGTP and 8-oxoGTP and therefore excludes 8-oxoguanine from DNA and RNA. The steady state levels of 8-oxoguanine in DNA of several organs including the brain are significantly reduced in hMTH1-Tg over-expressing mice. Conversely, MTH1-null mice exhibit a significantly higher level of 8-oxo-dGTP accumulation than that of the wild type.[9] Over-expression of hMTH1 prevents the age-dependent accumulation of DNA 8-oxoguanine that occurs in wild-type mice. The lower levels of oxidized guanines are associated with greater longevity. The hMTH1-Tg animals have a significantly longer lifespan than their wild-type littermates. These findings provide a link between ageing and oxidative DNA damage[8] (see DNA damage theory of aging).
Cancer
[edit]Studies have suggested that this enzyme plays a role in both preventing the formation of cancer cells and the proliferation of cancer cells. This makes it a topic of interest in cancer research, both as a potential method for healthy cells to prevent cancer and a weakness to target within existing cancer cells.
Eliminating the MTH1 gene in mice results in over three times more mice developing tumors compared to a control group.[10] The enzyme's much-studied ability to sanitize a cell's nucleotide pool prevents it from developing mutations, including cancerous ones. Specifically, another study found that MTH1 inhibition in cancer cells leads to incorporation of 8-oxo-dGTP and other oxidatively damaged nucleotides into the cell's DNA, damaging it and causing cell death.[11] However, cancer cells have also been shown to benefit from use of MTH1. Cells from malignant breast tumors exhibit extreme MTH1 expression compared to other human cells.[12] Because a cancer cell divides much more rapidly than a normal human cell, it is far more in need of an enzyme like MTH1 that prevents fatal mutations during replication. This property of cancer cells could allow for monitoring of cancer treatment efficacy by measuring MTH1 expression. Development of suitable probes for this purpose is currently underway.[13][14]
Disagreement exists concerning MTH1's functionality relative to prevention of DNA damage and cancer. Subsequent studies have had difficulty reproducing previously reported cytotoxic or antiproliferation effects of MTH1 inhibition on cancer cells, even calling into question whether MTH1 truly does serve to remove oxidatively damaged nucleotides from a cell's nucleotide pool.[15][16] One study of newly discovered MTH1 inhibitors suggests that these anticancer properties exhibited by the older MTH1 inhibitors may be due to off-target cytotoxic effects.[17] After revisiting the experiment, the original authors of this claim found that while the original MTH1 inhibitors in question lead to damaged nucleotides being incorporated into DNA, they demonstrate the others that do not induce toxicity fail to introduce the DNA lesion.[18] Research into this topic is ongoing.
As a drug target
[edit]MTH1 is a potential drug target to treat cancer, however there are conflicting results regarding the cytotoxicity of MTH1 inhibitors toward cancer cells.[19]
Karonudib, an MTH1 inhibitor, is currently being evaluated a phase I clinical trial for safety and tolerability.[18][20][21]
A potent and selective MTH1 inhibitor AZ13792138 has been developed by AstraZeneca has been made available as a chemical probe to academic researchers.[22] However AstraZeneca has found that neither AZ13792138 nor genetic knockdown of MTH1 displays any significant cytotoxicity to cancer cells.[23][24]
See also
[edit]References
[edit]- ^ Ponnambalam S, Jackson AP, LeBeau MM, Pravtcheva D, Ruddle FH, Alibert C, Parham P (December 1994). "Chromosomal location and some structural features of human clathrin light-chain genes (CLTA and CLTB)". Genomics. 24 (3): 440–4. doi:10.1006/geno.1994.1650. PMID 7713494.
- ^ Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M (November 1993). "Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis". The Journal of Biological Chemistry. 268 (31): 23524–30. doi:10.1016/S0021-9258(19)49494-5. PMID 8226881.
- ^ a b "Entrez Gene: NUDT1 nudix (nucleoside diphosphate linked moiety X)-type motif 1".
- ^ Rudling, Axel; Gustafsson, Robert; Almlöf, Ingrid; Homan, Evert; Scobie, Martin; Warpman Berglund, Ulrika; Helleday, Thomas; Stenmark, Pål; Carlsson, Jens (2017-10-12). "Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space". Journal of Medicinal Chemistry. 60 (19): 8160–8169. doi:10.1021/acs.jmedchem.7b01006. ISSN 1520-4804. PMID 28929756.
- ^ Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S (June 1991). "8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity". Proceedings of the National Academy of Sciences of the United States of America. 88 (11): 4690–4. Bibcode:1991PNAS...88.4690T. doi:10.1073/pnas.88.11.4690. PMC 51731. PMID 2052552.
- ^ a b Waz S, Nakamura T, Hirata K, Koga-Ogawa Y, Chirifu M, Arimori T, Tamada T, Ikemizu S, Nakabeppu Y, Yamagata Y (February 2017). "Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity". The Journal of Biological Chemistry. 292 (7): 2785–2794. doi:10.1074/jbc.m116.749713. PMC 5314174. PMID 28035004.
- ^ Yoshimura K, Ogawa T, Tsujimura M, Ishikawa K, Shigeoka S (September 2014). "Ectopic expression of the human MutT-type Nudix hydrolase, hMTH1, confers enhanced tolerance to oxidative stress in arabidopsis". Plant & Cell Physiology. 55 (9): 1534–43. doi:10.1093/pcp/pcu083. PMID 24928220.
- ^ a b De Luca G, Ventura I, Sanghez V, Russo MT, Ajmone-Cat MA, Cacci E, Martire A, Popoli P, Falcone G, Michelini F, Crescenzi M, Degan P, Minghetti L, Bignami M, Calamandrei G (August 2013). "Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1". Aging Cell. 12 (4): 695–705. doi:10.1111/acel.12094. PMID 23648059.
- ^ Kajitani K, Yamaguchi H, Dan Y, Furuichi M, Kang D, Nakabeppu Y (February 2006). "MTH1, an oxidized purine nucleoside triphosphatase, suppresses the accumulation of oxidative damage of nucleic acids in the hippocampal microglia during kainate-induced excitotoxicity". The Journal of Neuroscience. 26 (6): 1688–98. doi:10.1523/jneurosci.4948-05.2006. PMC 6793619. PMID 16467516.
- ^ Tsuzuki T, Egashira A, Igarashi H, Iwakuma T, Nakatsuru Y, Tominaga Y, Kawate H, Nakao K, Nakamura K, Ide F, Kura S, Nakabeppu Y, Katsuki M, Ishikawa T, Sekiguchi M (September 2001). "Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase". Proceedings of the National Academy of Sciences of the United States of America. 98 (20): 11456–61. Bibcode:2001PNAS...9811456T. doi:10.1073/pnas.191086798. PMC 58751. PMID 11572992.
- ^ Gad H, Koolmeister T, Jemth AS, Eshtad S, Jacques SA, Ström CE, et al. (April 2014). "MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool". Nature. 508 (7495): 215–21. Bibcode:2014Natur.508..215G. doi:10.1038/nature13181. PMID 24695224. S2CID 4468358.
- ^ Coskun E, Jaruga P, Jemth AS, Loseva O, Scanlan LD, Tona A, Lowenthal MS, Helleday T, Dizdaroglu M (September 2015). "Addiction to MTH1 protein results in intense expression in human breast cancer tissue as measured by liquid chromatography-isotope-dilution tandem mass spectrometry". DNA Repair. 33: 101–10. doi:10.1016/j.dnarep.2015.05.008. PMID 26202347.
- ^ Ji D, Beharry AA, Ford JM, Kool ET (July 2016). "A Chimeric ATP-Linked Nucleotide Enables Luminescence Signaling of Damage Surveillance by MTH1, a Cancer Target". Journal of the American Chemical Society. 138 (29): 9005–8. doi:10.1021/jacs.6b02895. PMC 5500214. PMID 27413803.
- ^ Samaranayake GJ, Troccoli CI, Huynh MQ, Win A, Ji D, Kool ET, Rai P (July 2017). "Towards a better understanding of MTH1 as a therapeutic target in RAS-driven cancer [abstract]". Cancer Res. 77 (13): 5473. doi:10.1158/1538-7445.AM2017-5473.
- ^ Petrocchi A, Leo E, Reyna NJ, Hamilton MM, Shi X, Parker CA, Mseeh F, Bardenhagen JP, Leonard P, Cross JB, Huang S, Jiang Y, Cardozo M, Draetta G, Marszalek JR, Toniatti C, Jones P, Lewis RT (March 2016). "Identification of potent and selective MTH1 inhibitors". Bioorganic & Medicinal Chemistry Letters. 26 (6): 1503–7. doi:10.1016/j.bmcl.2016.02.026. PMID 26898335.
- ^ Ellermann M, Eheim A, Rahm F, Viklund J, Guenther J, Andersson M, et al. (July 2017). "Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target". ACS Chemical Biology. 12 (8): 1986–1992. doi:10.1021/acschembio.7b00370. PMID 28679043.
- ^ Kettle JG, Alwan H, Bista M, Breed J, Davies NL, Eckersley K, Fillery S, Foote KM, Goodwin L, Jones DR, Käck H, Lau A, Nissink JW, Read J, Scott JS, Taylor B, Walker G, Wissler L, Wylot M (March 2016). "Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival". Journal of Medicinal Chemistry. 59 (6): 2346–61. doi:10.1021/acs.jmedchem.5b01760. PMID 26878898.
- ^ a b Warpman Berglund U, Sanjiv K, Gad H, Kalderén C, Koolmeister T, Pham T, et al. (December 2016). "Validation and development of MTH1 inhibitors for treatment of cancer". Annals of Oncology. 27 (12): 2275–2283. doi:10.1093/annonc/mdw429. PMID 27827301.
- ^ Samaranayake GJ, Huynh M, Rai P (2017). "MTH1 as a Chemotherapeutic Target: The Elephant in the Room". Cancers. 9 (5): 47. doi:10.3390/cancers9050047. PMC 5447957. PMID 28481306.
- ^ Thomas A (21 September 2016). "The Helleday Team presents potent MTH1 inhibitor 'Karonudib' and experimentally addresses the non-active MTH1 inhibitors". Helleday Laboratory. Retrieved 19 July 2017.
- ^ "MTH1, A Phase I, Study on Tumors Inhibition, First in Human, First in Class (MASTIFF)". ClinicalTrials.gov. U.S. National Institutes of Health. 14 February 2017. Retrieved 19 July 2017.
- ^ "AZ13792138". Open Innovation. AstraZeneca. 2016. Retrieved 19 July 2017.
- ^ Kettle JG, Alwan H, Bista M, Breed J, Davies NL, Eckersley K, et al. (2016). "Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival". Journal of Medicinal Chemistry. 59 (6): 2346–61. doi:10.1021/acs.jmedchem.5b01760. PMID 26878898.
- ^ Papeo G (2016). "MutT Homolog 1 (MTH1): The Silencing of a Target". Journal of Medicinal Chemistry. 59 (6): 2343–5. doi:10.1021/acs.jmedchem.6b00283. PMID 26924380.
Further reading
[edit]- Furuichi M, Yoshida MC, Oda H, Tajiri T, Nakabeppu Y, Tsuzuki T, Sekiguchi M (December 1994). "Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion". Genomics. 24 (3): 485–90. doi:10.1006/geno.1994.1657. PMID 7713500.
- Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K (June 1995). "Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria". The Journal of Biological Chemistry. 270 (24): 14659–65. doi:10.1074/jbc.270.24.14659. PMID 7782328.
- Oda H, Nakabeppu Y, Furuichi M, Sekiguchi M (July 1997). "Regulation of expression of the human MTH1 gene encoding 8-oxo-dGTPase. Alternative splicing of transcription products". The Journal of Biological Chemistry. 272 (28): 17843–50. doi:10.1074/jbc.272.28.17843. PMID 9211940.
- Oda H, Taketomi A, Maruyama R, Itoh R, Nishioka K, Yakushiji H, Suzuki T, Sekiguchi M, Nakabeppu Y (November 1999). "Multi-forms of human MTH1 polypeptides produced by alternative translation initiation and single nucleotide polymorphism". Nucleic Acids Research. 27 (22): 4335–43. doi:10.1093/nar/27.22.4335. PMC 148714. PMID 10536140.
- Sakai Y, Furuichi M, Takahashi M, Mishima M, Iwai S, Shirakawa M, Nakabeppu Y (March 2002). "A molecular basis for the selective recognition of 2-hydroxy-dATP and 8-oxo-dGTP by human MTH1". The Journal of Biological Chemistry. 277 (10): 8579–87. doi:10.1074/jbc.M110566200. PMID 11756418.
- Fujikawa K, Yakushiji H, Nakabeppu Y, Suzuki T, Masuda M, Ohshima H, Kasai H (February 2002). "8-Chloro-dGTP, a hypochlorous acid-modified nucleotide, is hydrolyzed by hMTH1, the human MutT homolog". FEBS Letters. 512 (1–3): 149–51. doi:10.1016/S0014-5793(02)02240-8. PMID 11852070.
- Topp H, Armbrust S, Lengger C, Schöch G, Davies J, Stichler W, Manz F, Fusch C (June 2002). "Renal excretion of 8-oxo-7,8-dihydro-2(')-deoxyguanosine: degradation rates of RNA and metabolic rate in humans". Archives of Biochemistry and Biophysics. 402 (1): 31–7. doi:10.1016/S0003-9861(02)00034-6. PMID 12051680.
- Takahashi M, Maraboeuf F, Sakai Y, Yakushiji H, Mishima M, Shirakawa M, Iwai S, Hayakawa H, Sekiguchi M, Nakabeppu Y (May 2002). "Role of tryptophan residues in the recognition of mutagenic oxidized nucleotides by human antimutator MTH1 protein". Journal of Molecular Biology. 319 (1): 129–39. doi:10.1016/S0022-2836(02)00163-8. PMID 12051941.
- Liu Z, Wang LE, Strom SS, Spitz MR, Babaian RJ, DiGiovanni J, Wei Q (March 2003). "Overexpression of hMTH in peripheral lymphocytes and risk of prostate cancer: a case-control analysis". Molecular Carcinogenesis. 36 (3): 123–9. doi:10.1002/mc.10108. PMID 12619034. S2CID 26364277.
- Ishibashi T, Hayakawa H, Sekiguchi M (May 2003). "A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides". EMBO Reports. 4 (5): 479–83. doi:10.1038/sj.embor.embor838. PMC 1319193. PMID 12717453.
- Kennedy CH, Pass HI, Mitchell JB (June 2003). "Expression of human MutT homologue (hMTH1) protein in primary non-small-cell lung carcinomas and histologically normal surrounding tissue". Free Radical Biology & Medicine. 34 (11): 1447–57. doi:10.1016/S0891-5849(03)00176-X. PMID 12757855.
- Yoshimura D, Sakumi K, Ohno M, Sakai Y, Furuichi M, Iwai S, Nakabeppu Y (September 2003). "An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress". The Journal of Biological Chemistry. 278 (39): 37965–73. doi:10.1074/jbc.M306201200. PMID 12857738.
- Kamiya H, Dugué L, Yakushiji H, Pochet S, Nakabeppu Y, Harashima H (2003). "Substrate recognition by the human MTH1 protein". Nucleic Acids Research. Supplement. 2 (2): 85–6. doi:10.1093/nass/2.1.85. PMID 12903117.
- Bialkowski K, Kasprzak KS (September 2003). "Inhibition of 8-oxo-2'-deoxyguanosine 5'-triphosphate pyrophosphohydrolase (8-oxo-dGTPase) activity of the antimutagenic human MTH1 protein by nucleoside 5'-diphosphates". Free Radical Biology & Medicine. 35 (6): 595–602. doi:10.1016/S0891-5849(03)00362-9. PMID 12957652.
- Kamiya H, Yakushiji H, Dugué L, Tanimoto M, Pochet S, Nakabeppu Y, Harashima H (February 2004). "Probing the substrate recognition mechanism of the human MTH1 protein by nucleotide analogs". Journal of Molecular Biology. 336 (4): 843–50. doi:10.1016/j.jmb.2003.12.060. PMID 15095864.
- Lowe, Derek (7 July 2017). "MTH1: From Hot Topic to Devalidation?". Science Translational Medicine. American Association for the Advancement of Science. Archived from the original on 10 July 2017. Retrieved 19 July 2017.
- Workman, Paul (10 July 2017). "Call to bioscientists: choose and use your chemicaI probes very carefully". The Institute of Cancer Research, London. The Institute of Cancer Research. Retrieved 19 July 2017.
External links
[edit]- 2-hydroxy-dATP+diphosphatase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

