Phosphoribulokinase
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
| phosphoribulokinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 3D cartoon depiction of a phosphoribulokinase protomer from Methanospirillum hungatei | |||||||||
| Identifiers | |||||||||
| EC no. | 2.7.1.19 | ||||||||
| CAS no. | 9030-60-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Phosphoribulokinase (PRK) (EC 2.7.1.19) is an essential photosynthetic enzyme that catalyzes the ATP-dependent phosphorylation of ribulose 5-phosphate (RuP) into ribulose 1,5-bisphosphate (RuBP), both intermediates in the Calvin Cycle. Its main function is to regenerate RuBP, which is the initial substrate and CO2-acceptor molecule of the Calvin Cycle.[1] PRK belongs to the family of transferase enzymes, specifically those transferring phosphorus-containing groups (phosphotransferases) to an alcohol group acceptor. Along with ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo), phosphoribulokinase is unique to the Calvin Cycle.[2] Therefore, PRK activity often determines the metabolic rate in organisms for which carbon fixation is key to survival.[3] Much initial work on PRK was done with spinach leaf extracts in the 1950s; subsequent studies of PRK in other photosynthetic prokaryotic and eukaryotic organisms have followed. The possibility that PRK might exist was first recognized by Weissbach et al. in 1954; for example, the group noted that carbon dioxide fixation in crude spinach extracts was enhanced by the addition of ATP.[3][4] The first purification of PRK was conducted by Hurwitz and colleagues in 1956.[5][6][7]
ATP + Mg2+ - D-ribulose 5-phosphate ADP + D-ribulose 1,5-bisphosphate 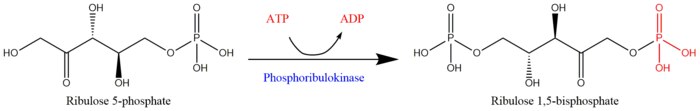
Reaction scheme for the regeneration of ribulose 1,5-bisphosphate from ribulose 5-phosphate by phosphoribulokinase[1]
The two substrates of PRK are ATP and D-ribulose 5-phosphate, whereas its two products are ADP and D-ribulose 1,5-bisphosphate. PRK activity requires the presence of a divalent metal cation like Mg2+, as indicated in the reaction above.[3]
Structure[edit]
The structure of PRK is different in prokaryotes and eukaryotes. Prokaryotic PRK's typically exist as octamers of 32 kDa subunits, while eukaryotic PRK's are often dimers of 40 kDa subunits.[8][9] Structural determinations for eukaryotic PRK have yet to be conducted, but prokaryotic PRK structures are still useful for rationalizing the regulation and mechanism of PRK. As of 2018, only two crystal structures have been resolved for this class of enzymes in Rhodobacter sphaeroides and Methanospirillum hungatei, with the respective PDB accession codes 1A7J and 5B3F.

Rhodobacter sphaeroides[edit]
In Rhodobacter sphaeroides, PRK (or RsPRK) exists as a homooctomer with protomers composed of seven-stranded mixed β-sheets, seven α-helices, and an auxiliary pair of anti-parallel β-strands.[10] The RsPRK subunit exhibits a protein folding analogous to the folding of nucleotide monophosphate (NMP) kinases.[3] Mutagenesis studies suggest that either Asp 42 or Asp 169 acts as the catalytic base that deprotonates the O1 hydroxyl oxygen on RuP for nucleophilic attack of ATP, while the other acts a ligand for a metal cation like Mg2+ (read mechanism below for more details).[10] Other residues present at the active site for RsPRK include His 45, Arg 49, Arg 168, and Arg 173, which are purportedly involved in RuP binding.[10] (See image at right).
Methanospirillum hungatei[edit]
In archaeal PRK of Methanospirillum hungatei, PRK (or MhPRK) exists as a homodimer of two protomers, each consisting of eight-stranded mixed β-sheets surrounded by α-helices and β-strands—similar to the structure of bacterial PRK from R. sphaeroides (see info. box above).[11] Although their quaternary structures differ and they have low amino acid sequence identity, MhPRK and RsPRK have structurally similar N-terminal domains as well as sequentially conserved residues like His 55, Lys 151, and Arg 154.[11]
Mechanism and Activity[edit]
PRK catalyzes the phosphorylation of RuP into RuBP. A catalytic residue in the enzyme (i.e. aspartate in RsPRK) deprotonates the O1 hydroxyl oxygen on RuP and activates it for nucleophilic attack of the γ-phosphoryl group of ATP.[10] As the γ-phosphoryl group is transferred from ATP to RuP, its stereochemistry inverts.[12] To allow for such inversion, the catalytic mechanism of PRK must not involve a phosphoryl-enzyme intermediate.[12]
Some studies suggest that both substrates (ATP and RuP) bind simultaneously to PRK and form a ternary complex. Others suggest that the substrate addition is sequential; the particular order by which substrates are added is still disputed, and may in fact, vary for different organisms.[13][14] In addition to binding its substrates, PRK also requires ligation to divalent metal cations like Mg2+ or Mn2+ for activity; Hg2+ has been demonstrated to inactivate the enzyme.[3][15]
Enzyme specificity[edit]
PRK shows high specificity for ribulose 5-phosphate. It does not act on any of the following substrates: D-xylulose 5-phosphate, fructose 6-phosphate, and sedoheptulose 7-phosphate.[15] However, at high concentrations, PRK may sometimes phosphorylate ribose 5-phosphate, a compound upstream the RuBP regeneration step in the Calvin Cycle.[15] Furthermore, PRK isolated from Alcaligenes eutrophus has been shown to use uridine triphosphate (UTP) and guanosine triphosphate (GTP) as alternative substrates to ATP.[8][3]
pH effects[edit]
The phosphorylation reaction proceeds with maximal velocity at pH 7.9, with no detectable activity at pH's below 5.5 or above 9.0.[15]
Regulation[edit]
The mechanisms by which prokaryotic and eukaryotic PRK's are regulated vary. Prokaryotic PRK's are typically subject to allosteric regulation while eukaryotic PRK's are often regulated by reversible thiol/disulfide exchange.[16] These differences are likely due to structural differences in their C-terminal domains[11]
Allosteric regulation of prokaryotic PRK[edit]
NADH is known to stimulate PRK activity, while AMP and phosphoenolpyruvate (PEP) are known to inhibit activity.[3] AMP has been shown to be involved in competitive inhibition in Thiobacillus ferrooxidans PRK.[17] On the other hand, PEP acts as a non-competitive inhibitor of PRK.[18]
Regulation of eukaryotic PRK[edit]
Eukaryotic PRK is typically regulated through the reversible oxidation/reduction of its cysteine sulfhydryl groups, but studies suggests that its activity can be regulated by other proteins or metabolites in the chloroplast. Of such metabolites, 6-phosphogluconate has been shown to be the most effective inhibitor of eukaryotic PRK by competing with RuP for the enzyme's active site.[19] This phenomenon may arise from the similarity in molecular structure between 6-phosphogluconate and RuP.
More recent work on the regulation of eukaryotic PRK has focused on its ability to form multi-enzyme complexes with other Calvin cycle enzymes such as glyceraldehyde 3-phosphate dehydrogenase (G3PDH) or RuBisCo.[20] In Chlamydomonas reinhardtii, chloroplast PRK and G3PDH exist as a bi-enzyme complex of 2 molecules of dimeric PRK and 2 molecules of tetrameric G3PDH thorough association by an Arg 64 residue, which may potentially transfer information between the two enzymes as well.[21]
Multi-enzyme complexes are likely to have more intricate regulatory mechanisms, and studies have already probed such processes. For example, it has been shown that PRK-glyceraldehyde 3-phosphate dehydrogenase complexes in Scenedesmus obliquus only dissociate to release activated forms of its constituent enzymes in the presence of NADPH, dithiothreitol (DTT), and thioredoxin.[22] Another topic of interest has been to compare the relative levels of PRK activity for when it is complexed to when it is not. For different photosynthetic eukaryotes, the enzyme activity of complexed PRK may be enhanced as opposed to free PRK, and vice versa.[23][24]
Other names[edit]
The systematic name of this enzyme class is ATP:D-ribulose-5-phosphate 1-phosphotransferase. Other names in common use include phosphopentokinase, ribulose-5-phosphate kinase, phosphopentokinase, phosphoribulokinase (phosphorylating), 5-phosphoribulose kinase, ribulose phosphate kinase, PKK, PRuK, and PRK.
References[edit]
- ^ a b Berg, Jeremy M.; Tymoczko, John L.; Gatto, Jr., Gregory J.; Stryer, Lubert (2015-04-08). Biochemistry (Eighth ed.). New York. ISBN 978-1464126109. OCLC 913469736.
{{cite book}}: CS1 maint: location missing publisher (link) - ^ Marsden WJ (September 16, 1983). "Purification and Molecular and Catalytic Properties of Phosphoribulokinase from the Cyanobacterium Chlorogloeopsis fritschii". Journal of General Microbiology. 130 (4): 999–1006. doi:10.1099/00221287-130-4-999.
- ^ a b c d e f g Miziorko HM (2000). "Phosphoribulokinase: Current Perspectives on the Structure/Function Basis for Regulation and Catalysis". In Purich DL (ed.). Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology - and Related Areas of Molecular Biology. Vol. 74. John Wiley & Sons, Inc. pp. 95–127. doi:10.1002/9780470123201.ch3. ISBN 9780470123201. PMID 10800594.
- ^ Weissbach A, Smyrniotis PZ, Horecker BL (July 1954). "Pentose phosphate and CO2 fixation with spinach extracts". Journal of the American Chemical Society. 76 (13): 3611–3612. doi:10.1021/ja01642a090.
- ^ Hurwitz J, Weissbach A, Horecker BL, Smyrniotis PZ (February 1956). "Spinach phosphoribulokinase". The Journal of Biological Chemistry. 218 (2): 769–83. doi:10.1016/S0021-9258(18)65841-7. PMID 13295229.
- ^ Racker E (July 1957). "The reductive pentose phosphate cycle. I. Phosphoribulokinase and ribulose diphosphate carboxylase". Archives of Biochemistry and Biophysics. 69: 300–10. doi:10.1016/0003-9861(57)90496-4. PMID 13445203.
- ^ Jakoby WB, Brummond DO, Ochoa S (February 1956). "Formation of 3-phosphoglyceric acid by carbon dioxide fixation with spinach leaf enzymes". The Journal of Biological Chemistry. 218 (2): 811–22. doi:10.1016/S0021-9258(18)65844-2. PMID 13295232.
- ^ a b Siebert K, Schobert P, Bowien B (March 1981). "Purification, some catalytic and molecular properties of phosphoribulokinase from Alcaligenes eutrophus". Biochimica et Biophysica Acta (BBA) - Enzymology. 658 (1): 35–44. doi:10.1016/0005-2744(81)90247-3. PMID 6260209.
- ^ Buchanan, Bob B. (2003-11-28). "Role of Light in the Regulation of Chloroplast Enzymes". Annu. Rev. Plant Physiol. 31: 341–374. doi:10.1146/annurev.pp.31.060180.002013.
- ^ a b c d Harrison DH, Runquist JA, Holub A, Miziorko HM (April 1998). "The crystal structure of phosphoribulokinase from Rhodobacter sphaeroides reveals a fold similar to that of adenylate kinase". Biochemistry. 37 (15): 5074–85. doi:10.1021/bi972805y. PMID 9548738.
- ^ a b c Kono T, Mehrotra S, Endo C, Kizu N, Matusda M, Kimura H, Mizohata E, Inoue T, Hasunuma T, Yokota A, Matsumura H, Ashida H (January 2017). "A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea". Nature Communications. 8: 14007. Bibcode:2017NatCo...814007K. doi:10.1038/ncomms14007. PMC 5241800. PMID 28082747.
- ^ a b Miziorko HM, Eckstein F (November 1984). "The stereochemical course of the ribulose-5-phosphate kinase-catalyzed reaction". The Journal of Biological Chemistry. 259 (21): 13037–40. doi:10.1016/S0021-9258(18)90652-6. PMID 6490643.
- ^ Lebreton S, Gontero B, Avilan L, Ricard J (December 1997). "Information transfer in multienzyme complexes--1. Thermodynamics of conformational constraints and memory effects in the bienzyme glyceraldehyde-3-phosphate-dehydrogenase-phosphoribulokinase complex of Chlamydomonas reinhardtii chloroplasts". European Journal of Biochemistry. 250 (2): 286–95. doi:10.1111/j.1432-1033.1997.0286a.x. PMID 9428675.
- ^ Wadano A, Nishikawa K, Hirahashi T, Satoh R, Iwaki T (1998-04-01). "Reaction mechanism of phosphoribulokinase from a cyanobacterium, Synechococcus PCC7942". Photosynthesis Research. 56 (1): 27–33. doi:10.1023/A:1005979801741. S2CID 21409736.
- ^ a b c d Hurwitz J (1962). [28c] Phosphoribulokinase. Methods in Enzymology. Vol. 5. pp. 258–261. doi:10.1016/s0076-6879(62)05214-3. ISBN 9780121818050.
- ^ Tabita FR (September 1980). "Pyridine nucleotide control and subunit structure of phosphoribulokinase from photosynthetic bacteria". Journal of Bacteriology. 143 (3): 1275–80. doi:10.1128/JB.143.3.1275-1280.1980. PMC 294495. PMID 6251028.
- ^ Gale NL, Beck JV (September 1966). "Competitive inhibition of phosphoribulokinase by AMP". Biochemical and Biophysical Research Communications. 24 (5): 792–6. doi:10.1016/0006-291X(66)90396-2. PMID 5970515.
- ^ Ballard RW, MacElroy RD (August 1971). "Phosphoenolpyruvate, a new inhibitor of phosphoribulokinase in pseudomonas facilis". Biochemical and Biophysical Research Communications. 44 (3): 614–8. doi:10.1016/s0006-291x(71)80127-4. PMID 4330777.
- ^ Gardemann, A.; Stitt, M.; Heldt, H.W. (1983-01-13). "Control of CO2 fixation. Regulation of spinach ribulose-5-phosphate kinase by stromal metabolite levels". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 722 (1): 51–60. doi:10.1016/0005-2728(83)90156-1.
- ^ Müller, Bruno (1972-08-01). "A Labile CO2-Fixing Enzyme Complex in Spinach Chloroplasts". Zeitschrift für Naturforschung B. 27 (8): 925–932. doi:10.1515/znb-1972-0814.
- ^ Avilan L, Gontero B, Lebreton S, Ricard J (December 1997). "Information transfer in multienzyme complexes--2. The role of Arg64 of Chlamydomonas reinhardtii phosphoribulokinase in the information transfer between glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase". European Journal of Biochemistry. 250 (2): 296–302. doi:10.1111/j.1432-1033.1997.0296a.x. PMID 9428676.
- ^ Nicholson S, Easterby JS, Powls R (January 1987). "Properties of a multimeric protein complex from chloroplasts possessing potential activities of NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase". European Journal of Biochemistry. 162 (2): 423–31. doi:10.1111/j.1432-1033.1987.tb10619.x. PMID 3026812.
- ^ Rault M, Gontero B, Ricard J (May 1991). "Thioredoxin activation of phosphoribulokinase in a chloroplast multi-enzyme complex". European Journal of Biochemistry. 197 (3): 791–7. doi:10.1111/j.1432-1033.1991.tb15973.x. PMID 1851485.
- ^ Gontero B, Mulliert G, Rault M, Giudici-Orticoni MT, Ricard J (November 1993). "Structural and functional properties of a multi-enzyme complex from spinach chloroplasts. 2. Modulation of the kinetic properties of enzymes in the aggregated state". European Journal of Biochemistry. 217 (3): 1075–82. doi:10.1111/j.1432-1033.1993.tb18339.x. PMID 8223631.

