Tibrovirus
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
| Tibrovirus | |
|---|---|
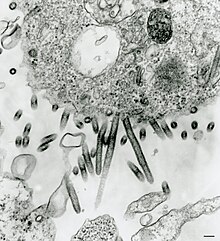 | |
| Sweetwater Branch tibrovirus (530 nm to 690 nm and up to 900 nm long, 65 nm to 75 nm in diameter)[2] | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Rhabdoviridae |
| Genus: | Tibrovirus |
| Species[1] | |
Tibrovirus is a poorly characterized genus of viruses in the family Rhabdoviridae, order Mononegavirales. There are 8 members of the genus.[3] Tibroviruses have been isolated from biting midges, cattle, and humans. None of the tibroviruses, except for Bas-Congo virus, have been associated with any diseases.
Genus members[edit]
- Bas-Congo virus (BASV) was discovered in 2009 in the Democratic Republic of Congo in a blood sample collected from a 32-year-old male who survived a severe illness resembling hemorrhagic fever.[4] BASV could not be isolated from the patient’s sample has not been established as a human pathogen.[5]
- Beatrice Hill virus (BHV) was isolated from a pool of biting midges (Culicoides peregrinus) in 1984 in Northern Territory, Australia.[6] BHV is poorly characterized and serological studies to assess its prevalence have not been carried out.
- Bivens arm virus (BAV) was isolated in 1981-1982 from a pool of biting midges in Florida. Anti-BAV antibodies have been detected in a variety of animals, including cattle, throughout Florida and the Caribbean.[7] There is no evidence of human infection or any disease associated with BAV.[7]
- Coastal Plains virus (CPV) was discovered in 1981 in Northern Territory, Australia in the blood of a healthy, asymptomatic steer.[8] No anti-CPV antibodies have ever been detected in humans and no disease has been associated with CPV.
- Ekpoma virus 1 (EKV-1) was discovered in 2015 in a blood sample collected from a healthy, 45-year-old woman living in Ekpoma, Nigeria.[9] EKV-1 was present in her blood at 4.5 million RNA copies/mL suggesting robust replication. Experiments using a recombinant vesicular stomatitis virus expressing the EKV-1 envelope glycoprotein indicate EKV-1 has very broad tropism and can efficiently enter nearly all types of human cells.[10] EKV-1 could not be isolated from the patient’s blood sample and live replication-competent virus is not available.
- Ekpoma virus 2 (EKV-2) was discovered in 2015 in a blood sample collected from a healthy, 19-year-old woman living in Ekpoma, Nigeria.[9] EKV-2 was present in her blood at 45,000 RNA copies/mL suggesting modest replication. Experiments using a recombinant vesicular stomatitis virus expressing the EKV-2 envelope glycoprotein indicate EKV-2 has very broad tropism similar to EKV-1.[10] EKV-2 also could not be isolated from the patient’s blood sample and live replication-competent virus is not available.
- Sweetwater branch virus (SWBV) was isolated along with BAV from pools of biting midges in Florida in 1981-1982. Like BAV, SWBV exposure in various animals is likely widespread. There is no evidence of human exposure or any disease associated with the virus.
- Tibrogargan virus (TIBV), the first tibrovirus discovered, was isolated from a pool of biting midges (Culicoides brevitarsis) in 1976 in Peachester, Australia.[11] TIBV appears to be widespread among cattle in Australia. A survey of more than 3,000 cattle found many herds were 100% seropositive. TIBV infection of humans has not been reported. TIBV is an orphan virus and not associated with any disease. Experimental infections of cattle produced viremia, but no observable signs of illness.[12]
Transmission[edit]
BHV, BAV, SWBV and TIBV were isolated from biting midges, suggesting that midges are the major arthropod vector for these viruses. It is not known how BASV, EKV-1 and EKV-2 are transmitted.[citation needed]
Genetic divergence[edit]
Tibroviruses are highly divergent. For example, overall amino acid homology among the human-associated tibroviruses (i.e. BASV, EKV-1 and EKV-2) ranges from 33% - 39%.[9]
Morphology[edit]
Tibrovirus virions are enveloped, but only the morphology of Tibrogargan virus and Sweetwater branch virus have been observed by electron microscopy.[2]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Tibrovirus | Bullet-shaped | Helical | Enveloped | Linear | Non-segmented |
Genome[edit]
Tibrovirus genomes are single-stranded, negative-sense RNA molecules approximately 13 kb in length. The genome encodes for the typical five proteins found in all rhabdoviruses: nucleocapsid (N), phosphoprotein (P), matrix (M), glycoprotein (G), and polymerase (L). However, there are three additional genes, U1-U3, that encode for proteins of unknown function.[13]
Life cycle[edit]
Viral replication is cytoplasmic. Entry into the host cell is achieved by attachment of the viral G glycoproteins to host receptors, which mediate clathrin-mediated endocytosis.[14][10] Replication follows the negative-stranded RNA virus replication model. Negative stranded RNA virus transcription, using polymerase stuttering is the method of transcription. The virus exits the host cell by budding, and tubule-guided viral movement.[citation needed]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Tibrovirus | Bovine | None | Clathrin-mediated endocytosis | Budding | Cytoplasm | Cytoplasm | Zoonosis; arthropod bite: midges |
References[edit]
- ^ "Virus Taxonomy: 2018b Release". International Committee on Taxonomy of Viruses (ICTV). March 2019. Retrieved 3 February 2020.
- ^ a b Popov VL, Tesh RB, Weaver SC, Vasilakis N. Electron Microscopy in Discovery of Novel and Emerging Viruses from the Collection of the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA). Viruses. 2019;11(5):477. Published 2019 May 25. doi:10.3390/v11050477
- ^ "International Committee on the Taxonomy of Viruses - Tibrovirus genus". Archived from the original on 8 August 2019.
- ^ Grard G, Fair JN, Lee D, et al. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa [published correction appears in PLoS Pathog. 2016 Mar;12(3):e1005503] [published correction appears in PLoS Pathog. 2017 Sep 7;13(9):e1006583]. PLoS Pathog. 2012;8(9):e1002924. doi:10.1371/journal.ppat.1002924
- ^ Branco, Luis M.; Garry, Robert F. (3 December 2018). "Bas-Congo virus - not an established pathogen". Retrieved 31 January 2020.
- ^ Standfast, H. A.; Dyce, A. L.; St George, T. D.; Muller, M. J.; Doherty, R. L.; Carley, J. G.; Filippich, C., Isolation of arboviruses from insects collected at Beatrice Hill, Northern Territory of Australia, 1974-1976. Aust J Biol Sci 1984, 37, (5-6), 351-66
- ^ a b Gibbs, E. P.; Calisher, C. H.; Tesh, R. B.; Lazuick, J. S.; Bowen, R.; Greiner, E. C., Bivens Arm virus: a new rhabdovirus isolated from Culicoides insignis in Florida and related to Tibrogargan virus of Australia. Vet Microbiol 1989, 19, (2), 141-50
- ^ Cybinski DH, Gard GP. Isolation of a new rhabdovirus in Australia related to Tibrogargan virus. Aust J Biol Sci. 1986;39(3):225–232. doi:10.1071/bi9860225
- ^ a b c Stremlau MH, Andersen KG, Folarin OA, et al. Discovery of novel rhabdoviruses in the blood of healthy individuals from West Africa. PLoS Negl Trop Dis. 2015;9(3):e0003631. Published 2015 Mar 17. doi:10.1371/journal.pntd.0003631
- ^ a b c Caì Y, Yú S, Jangra RK, et al. Human, Nonhuman Primate, and Bat Cells Are Broadly Susceptible to Tibrovirus Particle Cell Entry. Front Microbiol. 2019;10:856. Published 2019 Apr 26. doi:10.3389/fmicb.2019.00856
- ^ Cybinski, D. H.; St. George, T. D.; Standfast, H. A.; McGregor, A., Isolation of Tibrogargan virus, a new Australian rhabdovirus, from Culicoides brevitaris. Vet Microbiol 1980, 5, 301-308
- ^ Cybinski, D. H.; Gard, G. P., Isolation of a new rhabdovirus in Australia related to Tibrogargan virus. Aust J Biol Sci 1986, 39, (3), 225-32
- ^ "Viral Zone". ExPASy. Retrieved 13 August 2015.
- ^ Steffen I, Liss NM, Schneider BS, Fair JN, Chiu CY, Simmons G. Characterization of the Bas-Congo virus glycoprotein and its function in pseudotyped viruses. J Virol. 2013;87(17):9558–9568. doi:10.1128/JVI.01183-13