Demethylation
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
Demethylation is the chemical process resulting in the removal of a methyl group (CH3) from a molecule.[1][2] A common way of demethylation is the replacement of a methyl group by a hydrogen atom, resulting in a net loss of one carbon and two hydrogen atoms.
The counterpart of demethylation is methylation.
In biochemistry
[edit]
Dicamba, a widely used herbicide, biodegrades by demethylation to give 3,6-dichlorosalicylic acid, catalyzed by a dioxygenase enzyme.[3]
Demethylation is relevant to epigenetics. Demethylation of DNA is catalyzed by demethylases. These enzymes oxidize N-methyl groups, which occur in histones, in lysine derivatives, and in some forms of DNA.[4]
- R2N-CH3 + O → R2N-H + CH2O
One family of such oxidative enzymes is the cytochrome P450.[5] Alpha-ketoglutarate-dependent hydroxylases are also active for demethylation of DNA, operating by a similar stoichiometry.[6] These reactions, which proceed via hydroxylation, exploit the slightly weakened C-H bonds of methylamines and methyl ethers.
Demethylation of some sterols are steps in the biosynthesis of testosterone and cholesterol. Methyl groups are lost as formate.[7]
Biomass processing
[edit]Methoxy groups heavily decorate the biopolymer lignin. Much interest has been shown in converting this abundant form of biomass into useful chemicals (aside from fuel). One step in such processing is demethylation. [8][9] The demethylation of vanillin, a derivative of lignin, requires 250 °C (482 °F) and strong base.[10] Pulp and paper industry digests lignin using aqueous sodium sulfide, which partially depolymerizes the lignin. Delignification is accompanied by extensive O-demethylation, yielding methanethiol, which is emitted by paper mills.[11]
In organic chemistry
[edit]Demethylation often refers to cleavage of ethers, especially aryl ethers.[12]
Historically, aryl methyl ethers, including natural products such as codeine (O-methylmorphine), have been demethylated by heating the substance in molten pyridine hydrochloride (melting point 144 °C (291 °F)) at 180 to 220 °C (356 to 428 °F), sometimes with excess hydrogen chloride, in a process known as the Zeisel–Prey ether cleavage.[13][14] Quantitative analysis for aromatic methyl ethers can be performed by argentometric determination of the N-methylpyridinium chloride formed.[15] The mechanism of this reaction starts with proton transfer from pyridinium ion to the aryl methyl ether, a highly unfavorable step (K < 10−11) that accounts for the harsh conditions required, given the much weaker acidity of pyridinium (pKa = 5.2) compared to the protonated aryl methyl ether (an arylmethyloxonium ion, pKa = –6.7 for aryl = Ph[16]). This is followed by SN2 attack of the arylmethyloxonium ion at the methyl group by either pyridine or chloride ion (depending on the substrate) to give the free phenol and, ultimately, N-methylpyridinium chloride, either directly or by subsequent methyl transfer from methyl chloride to pyridine.[15]

Another classical (but, again, harsh) method for the removal of the methyl group of an aryl methyl ether is to heat the ether in a solution of hydrogen bromide or hydrogen iodide sometimes also with acetic acid.[17] The cleavage of ethers by hydrobromic or hydroiodic acid proceeds by protonation of the ether, followed by displacement by bromide or iodide. A slightly milder set of conditions uses cyclohexyl iodide (CyI, 10.0 equiv) in N,N-dimethylformamide to generate a small amount of hydrogen iodide in situ.[18]
Boron tribromide, which can be used at room temperature or below, is a more specialized reagent for the demethylation of aryl methyl ethers. The mechanism of ether dealkylation proceeds via the initial reversible formation of a Lewis acid-base adduct between the strongly Lewis acidic BBr3 and the Lewis basic ether. This Lewis adduct can reversibly dissociate to give a dibromoboryl oxonium cation and Br–. Rupture of the ether linkage occurs through the subsequent nucleophilic attack on the oxonium species by Br– to yield an aryloxydibromoborane and methyl bromide. Upon completion of the reaction, the phenol is liberated along with boric acid (H3BO3) and hydrobromic acid (aq. HBr) upon hydrolysis of the dibromoborane derivative during aqueous workup.[19]
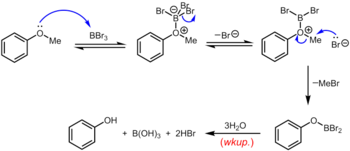
Stronger nucleophiles such as diorganophosphides (LiPPh2) also cleave aryl ethers, sometimes under mild conditions.[20] Other strong nucleophiles that have been employed include thiolate salts like EtSNa.[21]
Aromatic methyl ethers, particularly those with an adjacent carbonyl group, can be regioselectively demethylated using magnesium iodide etherate.[22] An example of this being used is in the synthesis of the natural product Calphostin A,[23] as seen below.

Methyl esters also are susceptible to demethylation, which is usually achieved by saponification. Highly specialized demethylations are abundant, such as the Krapcho decarboxylation:
A mixture of anethole, KOH, and alcohol was heated in an autoclave. Although the product of this reaction was the expected anol, a highly reactive dimerization product in the mother liquors called dianol was also discovered by Charles Dodds.
N-demethylation
[edit]N-demethylation of 3° amines is by the von Braun reaction, which uses BrCN as the reagent to give the corresponding nor- derivatives. A modern variation of the von Braun reaction was developed, where BrCN was superseded by ethyl chloroformate. The preparation of Paxil from arecoline is an application of this reaction, as well as the synthesis of GSK-372,475, for example.
The N-demethylation of imipramine gives desipramine.
See also
[edit]- Methylation, the addition of a methyl group to a substrate
References
[edit]- ^ Clayden, J.; Greeves, N.; Warren, S.; Wothers, P. (2001). Organic Chemistry. Oxford, Oxfordshire: Oxford University Press. ISBN 978-0-19-850346-0.
- ^ Smith, Michael B.; March, Jerry (2007), Advanced Organic Chemistry: Reactions, Mechanisms, and Structure (6th ed.), New York: Wiley-Interscience, ISBN 978-0-471-72091-1
- ^ Dumitru, Razvan; Jiang, Wen Zhi; Weeks, Donald P.; Wilson, Mark A. (2009). "Crystal Structure of Dicamba Monooxygenase: A Rieske Nonheme Oxygenase that Catalyzes Oxidative Demethylation". Journal of Molecular Biology. 392 (2): 498–510. doi:10.1016/j.jmb.2009.07.021. PMC 3109874. PMID 19616011.
- ^ Pedersen MT, Helin K (Nov 2010). "Histone demethylases in development and disease". Trends in Cell Biology. 20 (11): 662–71. doi:10.1016/j.tcb.2010.08.011. PMID 20863703.
- ^ Roland Sigel; Sigel, Astrid; Sigel, Helmut (2007). The Ubiquitous Roles of Cytochrome P450 Proteins: Metal Ions in Life Sciences. New York: Wiley. ISBN 978-0-470-01672-5.
- ^ Kohli RM, Zhang Y (October 2013). "TET enzymes, TDG and the dynamics of DNA demethylation". Nature. 502 (7472): 472–9. Bibcode:2013Natur.502..472K. doi:10.1038/nature12750. PMC 4046508. PMID 24153300.
- ^ Pietzke, Matthias; Meiser, Johannes; Vazquez, Alexei (2020). "Formate Metabolism in Health and Disease". Molecular Metabolism. 33: 23–37. doi:10.1016/j.molmet.2019.05.012. PMC 7056922. PMID 31402327.
- ^ Schmidt, Sandy (2022). "Expanding the repertoire of Rieske oxygenases for O-demethylations". Chem Catalysis. 2 (8): 1843–1845. doi:10.1016/j.checat.2022.07.005.
- ^ W. Boerjan; J. Ralph; M. Baucher (June 2003). "Lignin biosynthesis". Annu. Rev. Plant Biol. 54 (1): 519–549. doi:10.1146/annurev.arplant.54.031902.134938. PMID 14503002.
- ^ Irwin A. Pearl (1949). "Protocatechulic Acid". Organic Syntheses. 29: 85; Collected Volumes, vol. 3, p. 745.
- ^ Hansen, G. A. (1962). "Odor and Fallout Control in a Kraft Pulp Mill". Journal of the Air Pollution Control Association. 12 (9): 409–436. doi:10.1080/00022470.1962.10468107. PMID 13904415.
- ^ Weissman, Steven A.; Zewge, Daniel (2005). "Recent Advances in Ether Dealkylation". Tetrahedron. 61 (33): 7833–7863. doi:10.1016/j.tet.2005.05.041.
- ^ Lawson, J. A.; DeGraw, J. I. (1977). "An improved method for O-demethylation of codeine". Journal of Medicinal Chemistry. 20 (1): 165–166. doi:10.1021/jm00211a037. ISSN 0022-2623. PMID 833817.
- ^ Hassner, Alfred; Stumer, C. (2002). Organic syntheses based on name reactions (2nd ed.). Amsterdam: Pergamon. ISBN 9780080513348. OCLC 190810761.
- ^ a b Burwell, Robert L. (1954-08-01). "The Cleavage of Ethers". Chemical Reviews. 54 (4): 615–685. doi:10.1021/cr60170a003. ISSN 0009-2665.
- ^ Vollhardt, Peter; Schore, Neil (2014-01-01). Organic chemistry : structure and function (Seventh ed.). New York, NY. ISBN 9781464120275. OCLC 866584251.
{{cite book}}: CS1 maint: location missing publisher (link) - ^ Streitwieser, Andrew; Heathcock, Clayton H.; Kosower, Edward M. (1992). Introduction to organic chemistry (4th ed.). Upper Saddle River, N.J.: Prentice Hall. ISBN 978-0139738500. OCLC 52836313.
- ^ Zuo, Li; Yao, Shanyan; Wang, Wei; Duan, Wenhu (June 2008). "An efficient method for demethylation of aryl methyl ethers". Tetrahedron Letters. 49 (25): 4054–4056. doi:10.1016/j.tetlet.2008.04.070.
- ^ J. F. W. McOmie, D. E. West (1969). "3,3'-Dihydroxybiphenyl". Organic Syntheses. 49: 13; Collected Volumes, vol. 5, p. 412.
- ^ Robert E. Ireland; David M. Walba (1977). Organic Syntheses. Vol. 56. p. 44. doi:10.1002/0471264180.os056.11. ISBN 978-0471264224.
- ^ Mirrington, R. N.; Feutrill, G. I. (1973). "Orcinol Monomethyl Ether". Organic Syntheses. 53: 90. doi:10.15227/orgsyn.053.0090.
- ^ Yamaguchi, Seiji; Nedachi, Masahiro; Yokoyama, Hajime; Hirai, Yoshiro (October 1999). "Regioselective demethylation of 2,6-dimethoxybenzaldehydes with magnesium iodide etherate". Tetrahedron Letters. 40 (41): 7363–7365. doi:10.1016/S0040-4039(99)01411-2.
- ^ Merlic, Craig A.; Aldrich, Courtney C.; Albaneze-Walker, Jennifer; Saghatelian, Alan (1 April 2000). "Carbene Complexes in the Synthesis of Complex Natural Products: Total Synthesis of the Calphostins". Journal of the American Chemical Society. 122 (13): 3224–3225. doi:10.1021/ja994313+. PMC 3548573. PMID 23335811.

