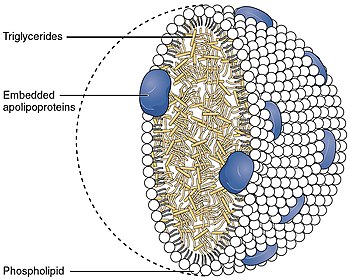
Apolipoprotein AI

Apolipoprotein AI (Apo-AI) is a protein that in humans is encoded by the APOA1 gene.[5][6] As the major component of HDL particles, it has a specific role in lipid metabolism.
Structure
[edit]APOA1 is located on chromosome 11, with its specific location being 11q23-q24. The gene contains 4 exons.[7] The encoded apolipoprotein AI, is a 28.1 kDa protein composed of 243 amino acids; 21 peptides have been observed through mass spectrometry data.[8][9] Due to alternative splicing, there exists multiple transcript variants of APOA1, including at least one which encodes a Apo-AI preprotein.[7]
Function
[edit]Apolipoprotein AI is the major protein component of high density lipoprotein (HDL) particles in plasma.[10]
Chylomicrons secreted from the intestinal enterocyte also contain Apo-AI, but it is quickly transferred to HDL in the bloodstream.[11]
The protein, as a component of HDL particles, enables efflux of fat molecules by accepting fats from within cells (including macrophages within the walls of arteries which have become overloaded with ingested fats from oxidized LDL particles) for transport (in the water outside cells) elsewhere, including back to LDL particles or to the liver for excretion.
It is a cofactor for lecithin–cholesterol acyltransferase (LCAT) which is responsible for the formation of most plasma cholesteryl esters. Apolipoprotein AI has also been isolated as a prostacyclin (PGI2) stabilizing factor, and thus may have an anticlotting effect.[12] Defects in the gene encoding it are associated with HDL deficiencies, including Tangier disease, and with systemic non-neuropathic amyloidosis.[7]
Apo-AI is often used as a biomarker for prediction of cardiovascular diseases. The ratio apoB-100/apoA-I (i.e. LDL & larger particles vs. HDL particles), NMR measured lipoprotein (low density lipoprotein (LDL)/(HDL) particle ratios even more so, has always had a stronger correlation with myocardial infarction event rates than older methods of measuring lipid transport in the water outside cells.[13]
Apo-AI is routinely measured using immunoassays such as ELISA or nephelometry.
Applications
[edit]Apo-AI can be used to create in vitro lipoprotein nanodiscs for cell-free membrane expression systems.[14]
Clinical significance
[edit]Activity associated with high HDL-C and protection from heart disease
[edit]As a major component of the high-density lipoprotein complex (protective "fat removal" particles), Apo-AI helps to clear fats, including cholesterol, from white blood cells within artery walls, making the white blood cells (WBCs) less likely to become fat overloaded, transform into foam cells, die and contribute to progressive atheroma. Five of nine men found to carry a mutation (E164X) who were at least 35 years of age had developed premature coronary artery disease.[15] One of four mutants of Apo-AI is present in roughly 0.3% of the Japanese population, but is found in 6% of those with low HDL cholesterol levels.[16]
ApoA-I Milano is a naturally occurring mutant of Apo-AI, found in a few families in Limone sul Garda, Italy, and, by genetic + church record family tree detective work, traced to a single individual, Giovanni Pomarelli, in the 18th century.[17] Described in 1980, it was the first known molecular abnormality of apolipoproteins.[18] Paradoxically, carriers of this mutation have very low HDL-C (HDL-Cholesterol) levels, but no increase in the risk of heart disease, often living to age 100 or older. This unusual observation was what lead Italian investigators to track down what was going on and lead to the discovery of apoA-I Milano (the city, Milano, ~160 km away, in which the researcher's lab was located). Biochemically, apo A1 contains an extra cysteine bridge, causing it to exist as a homodimer or as a heterodimer with Apo-AII. However, the enhanced cardioprotective activity of this mutant (which likely depends on fat & cholesterol efflux) cannot easily be replicated by other cysteine mutants.[19]
Recombinant Apo-AI Milano dimers formulated into liposomes can reduce atheromas in animal models by up to 30%.[20] Apo-AI Milano has also been shown in small clinical trials to have a statistically significant effect in reducing (reversing) plaque build-up on arterial walls.[21][22]
In human trials the reversal of plaque build-up was measured over the course of five weeks.[21][23]
Novel haplotypes within apolipoprotein AI-CIII-AIV gene cluster
[edit]A study from 2008 describes two novel susceptibility haplotypes, P2-S2-X1 and P1-S2-X1, discovered in ApoAI-CIII-AIV gene cluster on chromosome 11q23, which confer approximately threefold higher risk of coronary heart disease in normal[24] as well as in the patients having type 2 diabetes mellitus.[25]
Role in other diseases
[edit]A G/A polymorphism in the promoter of the APOA1 gene has been associated with the age at which Alzheimer disease is presented.[26] Protection from Alzheimer's disease by Apo-AI may rely on a synergistic interaction with alpha-tocopherol.[27] Amyloid deposited in the knee following surgery consists largely of Apo-AI secreted from chondrocytes (cartilage cells).[28] A wide variety of amyloidosis symptoms are associated with rare APOA1 mutants.
Apo-AI binds to lipopolysaccharide or endotoxin, and has a major role in the anti-endotoxin function of HDL.[29]
In one study, a decrease in Apo-AI levels was detected in schizophrenia patients' CSF, brain and peripheral tissues.[30]
Epistatic impact of Apo-AI
[edit]Apolipoprotein AI and ApoE interact epistatically to modulate triglyceride levels in coronary heart disease patients. Individually, neither Apo-AI nor ApoE was found to be associated with triglyceride (TG) levels, but pairwise epistasis (additive x additive model) explored their significant synergistic contributions with raised TG levels (P<0.01). [31]
Factors affecting Apo-AI activity
[edit]In a study from 2005 it was reported, that Apo-AI production is decreased by calcitriol. It was concluded, that this regulation happens on transcription level: calcitriol alters yet unknown coactivators or corepressors, resulting in repression of APOA1 promoter activity. Simultaneously, Apo-AI production was increased by vitamin D antagonist, ZK-191784.[32]
Exercise or statin treatment may cause an increase in HDL-C levels by inducing Apo-AI production, but this depends on the G/A promoter polymorphism.[33]
Interactions
[edit]Apolipoprotein A1 has been shown to interact with:
Potential binding partners
[edit]Apolipoprotein AI binding precursor, a relative of APOA-1 abbreviated APOA1BP, has a predicted biochemical interaction with carbohydrate kinase domain containing protein. The relationship between these two proteins is substantiated by cooccurance across genomes and coexpression.[37] The ortholog of CARKD in E. coli contains a domain not present in any eukaryotic ortholog. This domain has a high sequence identity to APOA1BP. CARKD is a protein of unknown function, and the biochemical basis for this interaction is unknown.
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000118137 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000032083 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Breslow JL, Ross D, McPherson J, Williams H, Kurnit D, Nussbaum AL, et al. (November 1982). "Isolation and characterization of cDNA clones for human apolipoprotein A-I". Proceedings of the National Academy of Sciences of the United States of America. 79 (22): 6861–6865. Bibcode:1982PNAS...79.6861B. doi:10.1073/pnas.79.22.6861. PMC 347233. PMID 6294659.
- ^ Arinami T, Hirano T, Kobayashi K, Yamanouchi Y, Hamaguchi H (June 1990). "Assignment of the apolipoprotein A-I gene to 11q23 based on RFLP in a case with a partial deletion of chromosome 11, del(11)(q23.3----qter)". Human Genetics. 85 (1): 39–40. doi:10.1007/BF00276323. PMID 1972696. S2CID 22613512.
- ^ a b c "Entrez Gene: APOA1 apolipoprotein A1".
- ^ Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, et al. (October 2013). "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research. 113 (9): 1043–1053. doi:10.1161/CIRCRESAHA.113.301151. PMC 4076475. PMID 23965338.
- ^ "Apolipoprotein A-IV". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB). Archived from the original on 5 March 2016. Retrieved 25 March 2015.
- ^ van der Vorst EP (2020). "High-Density Lipoproteins and Apolipoprotein A1". Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and other Body Fluid Proteins. Subcellular Biochemistry. Vol. 94. pp. 399–420. doi:10.1007/978-3-030-41769-7_16. ISBN 978-3-030-41768-0. PMID 32189309. S2CID 213180689.
- ^ Wasan KM, Brocks DR, Lee SD, Sachs-Barrable K, Thornton SJ (January 2008). "Impact of lipoproteins on the biological activity and disposition of hydrophobic drugs: implications for drug discovery". Nature Reviews. Drug Discovery. 7 (1): 84–99. doi:10.1038/nrd2353. PMID 18079757. S2CID 1989187.
- ^ Yui Y, Aoyama T, Morishita H, Takahashi M, Takatsu Y, Kawai C (September 1988). "Serum prostacyclin stabilizing factor is identical to apolipoprotein A-I (Apo A-I). A novel function of Apo A-I". The Journal of Clinical Investigation. 82 (3): 803–807. doi:10.1172/JCI113682. PMC 303586. PMID 3047170.
- ^ McQueen MJ, Hawken S, Wang X, Ounpuu S, Sniderman A, Probstfield J, et al. (July 2008). "Lipids, lipoproteins, and apolipoproteins as risk markers of myocardial infarction in 52 countries (the INTERHEART study): a case-control study". Lancet. 372 (9634): 224–233. doi:10.1016/S0140-6736(08)61076-4. PMID 18640459. S2CID 26567691.
- ^ Shelby ML, He W, Dang AT, Kuhl TL, Coleman MA (3 July 2019). "Cell-Free Co-Translational Approaches for Producing Mammalian Receptors: Expanding the Cell-Free Expression Toolbox Using Nanolipoproteins". Frontiers in Pharmacology. 10. Frontiers Media SA: 744. doi:10.3389/fphar.2019.00744. PMC 6616253. PMID 31333463.
- ^ Dastani Z, Dangoisse C, Boucher B, Desbiens K, Krimbou L, Dufour R, et al. (March 2006). "A novel nonsense apolipoprotein A-I mutation (apoA-I(E136X)) causes low HDL cholesterol in French Canadians". Atherosclerosis. 185 (1): 127–136. doi:10.1016/j.atherosclerosis.2005.05.028. PMID 16023124.
- ^ Yamakawa-Kobayashi K, Yanagi H, Fukayama H, Hirano C, Shimakura Y, Yamamoto N, et al. (February 1999). "Frequent occurrence of hypoalphalipoproteinemia due to mutant apolipoprotein A-I gene in the population: a population-based survey". Human Molecular Genetics. 8 (2): 331–336. doi:10.1093/hmg/8.2.331. PMID 9931341.
- ^ "The Long Saga of Apo-A1 Milano | in the Pipeline". 16 November 2016.
- ^ Franceschini G, Sirtori M, Gianfranceschi G, Sirtori CR (May 1981). "Relation between the HDL apoproteins and AI isoproteins in subjects with the AIMilano abnormality". Metabolism. 30 (5): 502–509. doi:10.1016/0026-0495(81)90188-8. PMID 6785551.
- ^ Zhu X, Wu G, Zeng W, Xue H, Chen B (June 2005). "Cysteine mutants of human apolipoprotein A-I: a study of secondary structural and functional properties". Journal of Lipid Research. 46 (6): 1303–1311. doi:10.1194/jlr.M400401-JLR200. PMID 15805548.
- ^ Chiesa G, Sirtori CR (April 2003). "Apolipoprotein A-I(Milano): current perspectives". Current Opinion in Lipidology. 14 (2): 159–163. doi:10.1097/00041433-200304000-00007. PMID 12642784. S2CID 75941726.
- ^ a b "Apo A-I-Milano Trial: Where are we now?". Cleveland Clinic. Retrieved 26 July 2008.
- ^ Nissen SE, Tsunoda T, Tuzcu EM, Schoenhagen P, Cooper CJ, Yasin M, et al. (November 2003). "Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial". JAMA. 290 (17): 2292–2300. doi:10.1001/jama.290.17.2292. PMID 14600188.
- ^ "Apo A-I Milano". Cedars-Sinai Heart Institute. Archived from the original on 21 December 2007. Retrieved 26 July 2008.
- ^ Singh P, Singh M, Kaur TP, Grewal SS (November 2008). "A novel haplotype in ApoAI-CIII-AIV gene region is detrimental to Northwest Indians with coronary heart disease". International Journal of Cardiology. 130 (3): e93–e95. doi:10.1016/j.ijcard.2007.07.029. PMID 17825930.
- ^ Singh P, Singh M, Gaur S, Kaur T (June 2007). "The ApoAI-CIII-AIV gene cluster and its relation to lipid levels in type 2 diabetes mellitus and coronary heart disease: determination of a novel susceptible haplotype". Diabetes & Vascular Disease Research. 4 (2): 124–129. doi:10.3132/dvdr.2007.030. PMID 17654446. S2CID 23793589.
- ^ Vollbach H, Heun R, Morris CM, Edwardson JA, McKeith IG, Jessen F, et al. (September 2005). "APOA1 polymorphism influences risk for early-onset nonfamiliar AD". Annals of Neurology. 58 (3): 436–441. doi:10.1002/ana.20593. PMID 16130094. S2CID 42148248.
- ^ Maezawa I, Jin LW, Woltjer RL, Maeda N, Martin GM, Montine TJ, et al. (December 2004). "Apolipoprotein E isoforms and apolipoprotein AI protect from amyloid precursor protein carboxy terminal fragment-associated cytotoxicity". Journal of Neurochemistry. 91 (6): 1312–1321. doi:10.1111/j.1471-4159.2004.02818.x. PMID 15584908. S2CID 30014992.
- ^ Solomon A, Murphy CL, Kestler D, Coriu D, Weiss DT, Makovitzky J, et al. (November 2006). "Amyloid contained in the knee joint meniscus is formed from apolipoprotein A-I". Arthritis and Rheumatism. 54 (11): 3545–3550. doi:10.1002/art.22201. PMID 17075859.
- ^ Ma J, Liao XL, Lou B, Wu MP (June 2004). "Role of apolipoprotein A-I in protecting against endotoxin toxicity". Acta Biochimica et Biophysica Sinica. 36 (6): 419–424. doi:10.1093/abbs/36.6.419. PMID 15188057.
- ^ Huang JT, Wang L, Prabakaran S, Wengenroth M, Lockstone HE, Koethe D, et al. (December 2008). "Independent protein-profiling studies show a decrease in apolipoprotein A1 levels in schizophrenia CSF, brain and peripheral tissues". Molecular Psychiatry. 13 (12): 1118–1128. doi:10.1038/sj.mp.4002108. PMID 17938634. S2CID 5576909.
- ^ Singh P, Singh M, Kaur T (May 2009). "Role of apolipoproteins E and A-I: epistatic villains of triglyceride mediation in coronary heart disease". International Journal of Cardiology. 134 (3): 410–412. doi:10.1016/j.ijcard.2007.12.102. PMID 18378026.
- ^ Wehmeier K, Beers A, Haas MJ, Wong NC, Steinmeyer A, Zugel U, et al. (October 2005). "Inhibition of apolipoprotein AI gene expression by 1, 25-dihydroxyvitamin D3". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1737 (1): 16–26. doi:10.1016/j.bbalip.2005.09.004. PMID 16236546.
- ^ Lahoz C, Peña R, Mostaza JM, Jiménez J, Subirats E, Pintó X, et al. (June 2003). "Apo A-I promoter polymorphism influences basal HDL-cholesterol and its response to pravastatin therapy". Atherosclerosis. 168 (2): 289–295. doi:10.1016/S0021-9150(03)00094-7. PMID 12801612.
- ^ Fitzgerald ML, Morris AL, Rhee JS, Andersson LP, Mendez AJ, Freeman MW (September 2002). "Naturally occurring mutations in the largest extracellular loops of ABCA1 can disrupt its direct interaction with apolipoprotein A-I". The Journal of Biological Chemistry. 277 (36): 33178–33187. doi:10.1074/jbc.M204996200. PMID 12084722.
- ^ Deeg MA, Bierman EL, Cheung MC (March 2001). "GPI-specific phospholipase D associates with an apoA-I- and apoA-IV-containing complex". Journal of Lipid Research. 42 (3): 442–451. doi:10.1016/S0022-2275(20)31669-2. PMID 11254757.
- ^ Pussinen PJ, Jauhiainen M, Metso J, Pyle LE, Marcel YL, Fidge NH, et al. (January 1998). "Binding of phospholipid transfer protein (PLTP) to apolipoproteins A-I and A-II: location of a PLTP binding domain in the amino terminal region of apoA-I". Journal of Lipid Research. 39 (1): 152–161. doi:10.1016/S0022-2275(20)34211-5. PMID 9469594.
- ^ "STRING: Known and Predicted Protein-Protein Interactions". Archived from the original on 18 July 2011.
External links
[edit]- Apolipoprotein+A-I at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Applied Research on Apolipoprotein-A1
- Human APOA1 genome location and APOA1 gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: P02647 (Human Apolipoprotein A-I) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: Q00623 (Mouse Apolipoprotein A-I) at the PDBe-KB.