Bat virome


The bat virome is the group of viruses associated with bats. Bats host a diverse array of viruses, including all seven types described by the Baltimore classification system: (I) double-stranded DNA viruses; (II) single-stranded DNA viruses; (III) double-stranded RNA viruses; (IV) positive-sense single-stranded RNA viruses; (V) negative-sense single-stranded RNA viruses; (VI) positive-sense single-stranded RNA viruses that replicate through a DNA intermediate; and (VII) double-stranded DNA viruses that replicate through a single-stranded RNA intermediate. The greatest share of bat-associated viruses identified as of 2020 are of type IV, in the family Coronaviridae.
Bats harbor several viruses that are zoonotic, or capable of infecting humans, and some bat-borne viruses are considered important emerging viruses.[1][2] These zoonotic viruses include the rabies virus, SARS-CoV, MERS-CoV, Marburg virus, Nipah virus, and Hendra virus. While research clearly indicates that SARS-CoV-2 originated in bats,[3] it is unknown how it was transmitted to humans, or if an intermediate host was involved. It has been speculated that bats may have a role in the ecology of the Ebola virus, though this is unconfirmed. While transmission of rabies from bats to humans usually occurs via biting, most other zoonotic bat viruses are transmitted by direct contact with infected bat fluids like urine, guano, or saliva, or through contact with an infected, non-bat intermediate host. There is no firm evidence that butchering or consuming bat meat can lead to viral transmission, though this has been speculated.
Despite the abundance of viruses associated with bats, they rarely become ill from viral infections, and rabies is the only viral illness known to kill bats. Much research has been conducted on bat virology, particularly bat immune response. Bats' immune systems differ from other mammals in their lack of several inflammasomes, which activate the body's inflammatory response, as well as a dampened stimulator of interferon genes (STING) response, which helps control host response to pathogens. Preliminary evidence indicates bats are thus more tolerant of infection than other mammals. While much research has centered on bats as a source of zoonotic disease, reviews have found mixed results on whether bats harbor more zoonotic viruses than other groups. A 2015 review found that bats do not harbor more zoonotic viruses than primates or rodents, though the three groups harbored more than other mammal orders.[4] In contrast, a 2020 review found that bats do not have more zoonotic viruses than any other bird or mammal group when viral diversity is measured relative to host diversity, as bats are the second-most diverse order of mammals.[5]
Viral diversity
[edit]| Virus family | No. sequences (n = 10,845) |
|---|---|
| Coronaviridae | |
| Rhabdoviridae | |
| Paramyxoviridae | |
| Astroviridae | |
| Adenoviridae | |
| Polyomaviridae | |
| Reoviridae | |
| Circoviridae | |
| Herpesviridae | |
| Flaviviridae | |
| Picornaviridae | |
| Parvoviridae | |
| Filoviridae | |
| Hepadnaviridae | |
| Papillomaviridae | |
| Hantaviridae | |
| Caliciviridae | |
| Peribunyaviridae | |
| Nairoviridae | |
| Retroviridae | |
| Orthomyxoviridae | |
| Phenuiviridae | |
| Poxviridae | |
| Picobirnaviridae | |
| Togaviridae | |
| Genomoviridae | |
| Bornaviridae | |
| Anelloviridae |
Viruses have been found in bat populations around the world. Bats harbor all groups of viruses in the Baltimore classification,[7] representing at least 28 families of viruses.[6] Most of the viruses harbored by bats are RNA viruses, though they are also known to have DNA viruses.[8] Bats are more tolerant of viruses than terrestrial mammals.[8] A single bat can host several different kinds of viruses without becoming ill.[9] Bats have also been shown to be more susceptible to reinfection with the same viruses, whereas other mammals, especially humans, have a greater propensity for developing varying degrees of immunity.[10][11] Their behavior and life history also make them "exquisitely suitable hosts of viruses and other disease agents", with long lifespans, the ability to enter torpor or hibernate, and their ability to traverse landscapes with daily and seasonal movement.[1]
Though bats harbor diverse viruses, they are rarely lethal to the bat host. Only the rabies virus and a few other lyssaviruses have been confirmed to kill bats.[7] Various factors have been implicated in bats' ability to survive viral infections. One possibility is bats' use of flight. Flight produces a fever-like response, resulting in elevated temperature (up to 38 °C (100 °F)) and metabolic rate. Additionally, this fever-like response may help them cope with actual fevers upon getting a viral infection.[7] Some research indicates that bats' immune systems have allowed them to cope with a variety of viruses. A 2018 study found that bats have a dampened STING response compared to other mammals, which could allow them to respond to viral threats without over-responding.[8] STING is a signaling molecule that helps coordinate various host defense genes against pathogens.[12] The authors of the study concluded that "the weakened, but not entirely lost, functionality of STING may have profound impact for bats to maintain the balanced state of 'effective response' but not 'over response' against viruses."[8]
Additionally, bats lack several inflammasomes found in other mammals;[8] other inflammasomes are present with a greatly reduced response.[13] While inflammation is an immune response to viruses, excessive inflammation is damaging to the body, and viruses like severe acute respiratory syndrome coronavirus (SARS-CoV) are known to kill humans by inducing excessive inflammation. Bats' immune systems may have evolved to be more tolerant of stressors such as viral infections compared to other mammals.[14]
Transmission to humans
[edit]
The vast majority of bat viruses have no zoonotic potential, meaning they cannot be transmitted to humans.[6] The zoonotic viruses have four possible routes of transmission to humans: contact with bat body fluids (blood, saliva, urine, feces); intermediate hosts; environmental exposure; and blood-feeding arthropods.[15] Lyssaviruses like the rabies virus are transmitted from bats to humans via biting. Transmission of most other viruses does not appear to take place via biting, however. Contact with bat fluids such as guano, urine, and saliva is an important source of spillover from bats to humans. Other mammals may play a role in transmitting bat viruses to people, with pig farms a source of bat-borne viruses in Malaysia and Australia.[15][16] Other possible transmission routes of bat-borne viruses are more speculative. It is possible but unconfirmed that hunting, butchering, and consuming bat meat can result in viral spillover. While arthropods like mosquitoes, ticks, and fleas may transmit viral infections from other mammals to humans, it is highly speculative that arthropods play a role in mediating bat viruses to humans. There is little evidence of environmental transmission of viruses from bats to humans, meaning that bat-borne virus do not persist in the environment for long. However, a limited number of studies have been conducted on the subject.[15]
Bats compared to other viral reservoirs
[edit]Bats and their viruses may be the subject of more research than viruses found in other mammal orders, an example of research bias. A 2015 review found that from 1999 to 2013, there were 300–1200 papers published about bat viruses annually, compared to 12–45 publications for marsupial viruses and only 1–9 studies for sloth viruses. The same review found that bats do not have significantly greater viral diversity than other mammal groups. Bats, rodents, and primates all harbored significantly more zoonotic viruses than other mammal groups, though the differences among the aforementioned three groups were not significant (bats have no more zoonotic viruses than rodents and primates).[4] A 2020 review of mammals and birds found that the identity of the taxonomic groups did not have any impact on the probability of harboring zoonotic viruses. Instead, more diverse groups had greater viral diversity. Bat life history traits and immunity, while likely influential in determining bat viral communities, were not associated with a greater probability of viral spillover into humans.[5]
Sampling
[edit]
Bats are sampled for viruses in a variety of ways. They can be tested for seropositivity for a given virus using a method like ELISA, which determines whether or not they have the corresponding antibodies for the virus. They can also be surveyed using molecular detection techniques like PCR (polymerase chain reaction), which can be used to replicate and amplify viral sequences. Histopathology, which is the microscopic examination of tissue, can also be used. Viruses have been isolated from bat blood, saliva, feces, tissue, and urine. Some sampling is non-invasive and does not require killing the bat for sampling, whereas other sampling requires sacrificing the animal first. A 2016 review found no significant difference in total number of viruses found and new viruses discovered between lethal and non-lethal studies. Several species of threatened bat have been killed for viral sampling, including the Comoro rousette, Hildegarde's tomb bat, Natal free-tailed bat, and the long-fingered bat.[17]
Double-stranded DNA viruses
[edit]Adenoviruses
[edit]Adenoviruses have been detected in bat guano, urine, and oral and rectal swabs. They have been found in both megabats and microbats across a large geographic area. Bat adenoviruses are closely related to those finds in canids.[18] The greatest diversity of bat adenoviruses has been found in Eurasia, though the virus family may be undersampled in bats overall.[7]
Herpesviruses
[edit]Diverse herpesviruses have been found in bats in North and South America, Asia, Africa, and Europe,[18] including representatives of the three subfamilies, alpha-, beta-, and gammaherpesviruses.[7] Bat-hosted herpesviruses include the species Pteropodid alphaherpesvirus 1 and Vespertilionid gammaherpesvirus 1.[19]
Papillomaviruses
[edit]Papillomaviruses were first detected in bats in 2006, in the Egyptian fruit bat. They have since been identified in several other bat species, including the serotine bat, greater horseshoe bat, and the straw-colored fruit bat. Five distinct lineages of bat papillomaviruses have been recognized.[18]
Single-stranded DNA viruses
[edit]Anelloviruses
[edit]No anellovirus is known to cause disease in humans.[7] The first bat anellovirus, a Torque teno virus, was found in a Mexican free-tailed bat.[20] Novel anelloviruses have also been detected in two leaf-nosed bat species: the common vampire bat and Seba's short-tailed bat. The bat anelloviruses and one opossum anellovirus have been included in the proposed genus Sigmatorquevirus.[21]
Circoviruses
[edit]Circoviruses, family Circoviridae, are among the most diverse of all viruses.[22] Like anelloviruses, circoviruses are not associated with any disease in humans.[7] About a third of all circoviruses are associated with bats, found in North and South America, Europe, and Asia.[22] A study of horseshoe and vesper bats in China identified circoviruses from the genera Circovirus and Cyclovirus.[23]
Parvoviruses
[edit]Several kinds of parvoviruses are considered important for human and animal health. Several strains of parvovirus have been identified from bat guano in the US states of Texas and California. Serum analysis of the straw-colored fruit bat and Jamaican fruit bat led to the identification of two new parvoviruses. Bat parvoviruses are in the subfamily Parvovirinae, closely resembling the genera Protoparvovirus, Erythrovirus, and Bocaparvovirus.[18]
Double-stranded RNA viruses
[edit]Reoviruses
[edit]| Virus name | Year identified | Host | Location |
|---|---|---|---|
| Nelson Bay virus | 1968 | Bat | Australia |
| Pulau virus | 1999 | Bat | Malaysia |
| Melaka virus | 2006 | Human | Malaysia |
| Kampar virus | 2006 | Human | Malaysia |
| HK23629/07 | 2007 | Human | Hong Kong |
| Miyazaki-Bali/2007 | 2007 | Human | Indonesia/Japan |
| Sikamat virus | 2010 | Human | Malaysia |
| Xi River virus | 2010 | Bat | China |
| Indonesia/2010 | 2010 | Bat | Indonesia/Italy |
Zoonotic
[edit]Some disease-causing reovirus species are associated with bats. One such virus is Melaka virus, which was linked to illness in a Malaysian man and his two children in 2006.[25][26] The man said that a bat had been in his home a week before he became ill, and the virus was closely related to other reoviruses linked to bats. Kampar virus was identified a few months later in another Malaysian man. Though he had no known contact with bats, Kampar virus is closely related to Melaka virus. Several other reovirus strains identified in ill humans are known as Miyazaki-Bali/2007, Sikamat virus, and SI-MRV01. No reoviruses linked to bats have caused death in humans.[25]
Other
[edit]Reoviruses include many viruses that do not cause disease in humans, including several found in bats. One reovirus species associated with bats is Nelson Bay orthoreovirus, sometimes called Pteropine orthoreovirus (PRV), which is an orthoreovirus; several virus strains of it have been identified in bats. The type member of Nelson Bay orthoreovirus is Nelson Bay virus (NBV), which was first identified in 1970 from the blood of a gray-headed flying fox in New South Wales, Australia. NBV was the first reovirus to be isolated from a bat species. Another strain of Nelson Bay orthoreovirus associated with bats is Pulau virus, which was first identified from the small flying fox of Tioman Island in 2006. Other viruses include Broome orthoreovirus from the little red flying fox of Broome, Western Australia; Xi River virus from Leschenault's rousette in Guangdong, China; and Cangyuan virus also from Leschenault's rousette.[25] Several mammalian orthoreoviruses are associated with bats, including at least three from Germany and 19 from Italy. These were found in pipistrelles, the brown long-eared bat, and the whiskered bat.[25]
Orbiviruses have been isolated from bats, including Ife virus from the straw-colored fruit bat, Japanaut virus from the common blossom bat, and Fomédé virus from Nycteris species.[25]
Positive-sense single-stranded RNA viruses
[edit]Astroviruses
[edit]Astroviruses have been found in several genera of bat in the Old World, including Miniopterus, Myotis, Hipposideros, Rhinolophus, Pipistrellus, Scotophilus, and Taphozous,[18] though none in Africa.[7] Bats have very high prevalence rates of astroviruses; studies in Hong Kong and mainland China found prevalence rates approaching 50% from anal swabs. No astroviruses identified in bats are associated with disease in humans.[18]
Caliciviruses
[edit]Bat caliciviruses were first identified in Hong Kong in the Pomona roundleaf bat,[18] and were later identified from tricolored bats in the US state of Maryland. Bat caliciviruses are similar to the genera Sapovirus and Valovirus, with noroviruses also detected from two microbat species in China.[27]
Coronaviruses
[edit]SARS-CoV, SARS-CoV-2, and MERS-CoV
[edit]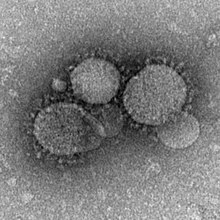
Several zoonotic coronaviruses are associated with bats, including severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome-related coronavirus (MERS-CoV).[28] Severe acute respiratory syndrome coronavirus 2 is another zoonotic coronavirus likely originating in bats.[29][30] SARS-CoV causes the disease severe acute respiratory syndrome (SARS) in humans. The first documented case of SARS was in November 2002 in Foshan, China.[28] It became an epidemic, affecting 28 countries around the world with 8,096 cases and 774 deaths.[28] The natural reservoir of SARS-CoV was identified as bats, with the Chinese rufous horseshoe bat considered a particularly strong candidate after a coronavirus was recovered from a colony that had 95% nucleotide sequence similarity to SARS-CoV.[28] There is uncertainty on whether or not animals like Himalayan palm civets and raccoon dogs were intermediate hosts that facilitated the spread of the virus from bats to humans, or if humans acquired the virus directly from bats.[28][31]
The first human case of Middle East respiratory syndrome (MERS) was in June 2012 in Jeddah, Saudi Arabia.[28] As of November 2019, 2,494 cases of MERS have been reported in twenty-seven countries, resulting in 858 fatalities.[32] It is believed that MERS-CoV originated in bats, though camels are likely the intermediate host through which humans became infected. Human-to-human transmission is possible, though does not easily occur.[33]
The COVID-19 pandemic in humans started in Wuhan, China in 2019.[34] Genetic analyses of SARS-COV-2 showed that it was highly similar to viruses found in horseshoe bats, with 96% similarity to a virus isolated from the intermediate horseshoe bat. Due to similarity with known bat coronaviruses, data "clearly indicates" that the natural reservoirs of SARS-COV-2 are bats. It is yet unclear how the virus was transmitted to humans, though an intermediate host may have been involved.[3] Phylogenetic reconstruction of SARS-CoV-2 suggests that the strain that caused a human pandemic diverged from the strain found in bats decades ago, likely between 1950 and 1980.[35]
Other
[edit]Bats harbor a great diversity of coronaviruses, with sampling by the EcoHealth Alliance in China alone identifying about 400 new strains of coronavirus.[36] A study of coronavirus diversity harbored by bats in eastern Thailand revealed forty-seven coronaviruses.[37]
Flaviviruses
[edit]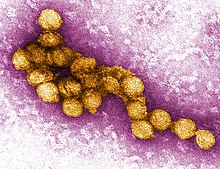
Most flaviviruses are transmitted via arthropods, but bats may play a role in the ecology of some species. Several strains of Dengue virus have been found in bats in the Americas, and West Nile virus has been identified in fruit bats in South India. Serological studies indicate that West Nile virus may also be present in bats in North America and the Yucatán Peninsula. Saint Louis encephalitis virus has been detected in bats in the US states of Texas and Ohio, as well as the Yucatán Peninsula. Japanese encephalitis virus or its associated antibodies have been found in several bat species throughout Asia. Other flaviviruses detected in bats include Sepik virus, Entebbe bat virus, Sokuluk virus, Yokose virus, Dakar bat virus, Bukalasa bat virus, Carey Island virus, Phnom Penh bat virus, Rio Bravo bat virus, Montana myotis leukoencephalitis virus, and Tamana bat virus.[18]
Picornaviruses
[edit]Several genera of picornaviruses have been found in bats, including Kobuvirus, Sapelovirus, Cardiovirus, and Senecavirus.[18] Picornaviruses have been identified from a diverse array of bat species around the world.[7]
Negative-sense single-stranded RNA viruses
[edit]Arenaviruses
[edit]Arenaviruses are mainly associated with rodents, though some can cause illness in humans. The first arenavirus identified in bats was Tacaribe mammarenavirus, which was isolated from Jamaican fruit bats and the great fruit-eating bat. Antibody response associated with Tacaribe virus has also been found in the common vampire bat, the little yellow-shouldered bat, and Heller's broad-nosed bat. It is unclear if bats are the natural reservoir of Tacaribe virus. There has been one known human infection by Tacaribe virus, though it was accidentally acquired in a laboratory setting.[18]
Hantaviruses
[edit]Hantaviruses, family Hantaviridae, naturally occur in vertebrates. All bat-associated hantaviruses are in the subfamily Mammantavirinae. Of the four genera within the subfamily, Loanvirus and Mobatvirus are the genera that have been documented in various bats. Almost all bat hantaviruses have been identified from microbats.[38] Mouyassue virus has been identified from the banana pipistrelle in Ivory Coast and the Cape serotine in Ethiopia;[38] Magboi virus from the hairy slit-faced bat in Sierra Leone; Xuan Son virus from the Pomona roundleaf bat in Vietnam; Huangpi virus from the Japanese house bat in China; Longquan loanvirus from several horseshoe bats in China;[18] Makokou virus from Noack's roundleaf bat in Gabon; Đakrông virus from Stoliczka's trident bat in Vietnam;[38] Brno loanvirus from the common noctule in the Czech Republic;[38] and Laibin mobatvirus from the black-bearded tomb bat in China.[39] As of 2019, only Quezon mobatvirus has been identified from a megabat, as it was identified from a Geoffroy's rousette in the Philippines.[38] Bat hantaviruses are not associated with illness in humans.[18][38]
Filoviruses
[edit]Marburgvirus and Ebolavirus
[edit]
Filoviridae is a family of virus containing two genera associated with bats: Marburgvirus and Ebolavirus, which contain the species that cause Marburg virus disease and Ebola virus disease, respectively. Though relatively few disease outbreaks are caused by filoviruses, they are of high concern due to their extreme virulence, or capacity to cause harm to their hosts. Filovirus outbreaks typically have high mortality rates in humans. Though the first filovirus was identified in 1967, it took more than twenty years to identify any natural reservoirs.[40]
Ebola virus disease is a relatively rare but life-threatening illness in humans, with an average mortality rate of 50% (though individual outbreaks may be as high as 90% mortality). The first outbreaks were in 1976 in South Sudan and Democratic Republic of the Congo.[41] The natural reservoirs of ebolaviruses are unknown.[42][43][44] However, some evidence indicates that megabats may be natural reservoirs.[40][41] Several megabat species have tested seropositive for antibodies against ebolaviruses, including the hammer-headed bat, Franquet's epauletted fruit bat, and little collared fruit bat.[40] Among others, it has been posited that the Western African Ebola virus epidemic began with a spillover event from an Angolan free-tailed bat to a human.[45] Other possible reservoirs include non-human primates,[42] rodents, shrews, carnivores, and ungulates.[46] Definitively stating that fruit bats are natural reservoirs is problematic; as of 2017, researchers have been largely unable to isolate ebolaviruses or their viral RNA sequences from fruit bats. Additionally, bats typically have low level of ebolavirus-associated antibodies, and seropositivity in bats is not strongly correlated to human outbreaks.[44]
Marburg virus disease (MVD) was first identified in 1967 during simultaneous outbreaks in Marburg and Frankfurt in Germany, and Belgrade, Serbia. MVD is highly virulent, with an average human mortality rate of 50%, but as high as 88% for individual outbreaks.[47] MVD is caused by Marburg virus and the closely related Ravn virus, which was formerly considered synonymous with Marburg virus.[48] Marburg virus was first detected in the Egyptian fruit bat in 2007,[40] which is now recognized as the natural reservoir of the virus.[47] Marburg virus has been detected in Egyptian fruit bats in Gabon, Democratic Republic of the Congo, Kenya, and Uganda.[40] Spillover from Egyptian fruit bats occurs when humans spend prolonged time in mines or caves inhabited by the bats,[47] though the exact mechanism of transmission is unclear.[40] Human-to-human transmission occurs through direct contact with infected bodily fluids, including blood or semen, or indirectly through contact with bedding or clothing exposed to these fluids.[47]
Other
[edit]Lloviu virus, a kind of filovirus in the genus Cuevavirus, has been identified from the common bent-wing bat in Spain.[40] Another filovirus, Bombali ebolavirus, has been isolated from free-tailed bats, including the little free-tailed bat and the Angolan free-tailed bat.[49] Neither Lloviu virus nor Bombali ebolavirus is associated with illness in humans.[50][49] Genomic RNA associated with Mengla dianlovirus, though not the virus itself, has been identified from Rousettus bats in China.[49]
Rhabdoviruses
[edit]Rabies-causing viruses
[edit]
Lyssaviruses (from the genus Lyssavirus in the family Rhabdoviridae) include the rabies virus, Australian bat lyssavirus, and other related viruses, many of which are also harbored by bats. Unlike most other viruses in the family Rhabdoviridae, which are transmitted by arthropods, lyssaviruses are transmitted by mammals, most frequently through biting. All mammals are susceptible to lyssaviruses, though bats and carnivores are the most common natural reservoirs. The vast majority of human rabies cases are a result of the rabies virus, with only twelve other human cases attributed to other lyssaviruses as of 2015.[51] These rarer lyssaviruses associated with bats include Duvenhage lyssavirus (three human cases as of 2015); European bat 1 lyssavirus (one human case as of 2015); European bat 2 lyssavirus (two human cases as of 2015); and Irkut lyssavirus (one human case as of 2015). Microbats are suspected as the reservoirs of these four uncommon lyssaviruses.[51][52]
After transmission has occurred, the average human is asymptomatic for two months, though the incubation period can be as short as a week or as long as several years.[51] Italian scientist Antonio Carini was the first to hypothesize that rabies virus could be transmitted by bats, which he did in 1911. This same conclusion was reached by Hélder Queiroz in 1934 and Joseph Lennox Pawan in 1936. Vampire bats were the first to be documented with rabies; in 1953, an insectivorous bat in Florida was discovered with rabies, making it the first documented occurrence in an insectivorous species outside the vampire bats' ranges.[53] Bats have an overall low prevalence of rabies virus, with a majority of surveys of apparently healthy individuals showing rabies incidence of 0.0–0.5%.[51] Sick bats are more likely to be submitted for rabies testing than apparently healthy bats, known as sampling bias,[54] with most studies reporting rabies incidence of 5–20% in sick or dead bats.[51] Rabies virus exposure can be fatal in bats, though it is likely that the majority of individuals do not develop the disease after exposure.[51] In non-bat mammals, exposure to the rabies virus almost always leads to death.[52]

Globally, dogs are by far the most common source of human rabies deaths.[55] Bats are the most common source of rabies in humans in North and South America, Western Europe, and Australia.[56] Many feeding guilds of bats may transmit rabies to humans, including insectivorous, frugivorous, nectarivorous, omnivorous, sanguivorous, and carnivorous species.[56] The common vampire bat is a source of human rabies in Central and South America, though the frequency at which humans are bitten is poorly understood.[57] Between 1993 and 2002, the majority of human rabies cases associated with bats in the Americas were the result of non-vampire bats.[52] In North America, about half of human rabies instances are cryptic, meaning that the patient has no known bite history.[51] While it has been speculated that rabies virus could be transmitted through aerosols, studies of the rabies virus have concluded that this is only feasible in limited conditions. These conditions include a very large colony of bats in a hot and humid cave with poor ventilation. While two human deaths in 1956 and 1959 had been tentatively attributed to aerosolization of the rabies virus after entering a cave with bats, "investigations of the 2 reported human cases revealed that both infections could be explained by means other than aerosol transmission".[58] It is instead generally thought that most instances of cryptic rabies are the result of an unknown bat bite.[51] Bites from a bat can be so small that they are not visible without magnification equipment, for example. Outside of bites, rabies virus exposure can also occur if infected fluids come in contact with a mucous membrane or a break in the skin.[58]
Other
[edit]Many bat lyssaviruses are not associated with infection in humans. These include Lagos bat lyssavirus, Shimoni bat lyssavirus, Khujand lyssavirus, Aravan lyssavirus, Bokeloh bat lyssavirus, West Caucasian bat lyssavirus, and Lleida bat lyssavirus.[52][51] Lagos bat lyssavirus, also known as Lagos bat virus (LBV), has been isolated from a megabat in sub-Saharan Africa.[51] This lyssavirus has four distinct lineages, all of which are found in the straw-colored fruit bat.[59]
Rhabdoviruses from other genera have been identified in bats. This includes several from the genus Ledantevirus: Kern Canyon virus, which was found in the Yuma myotis in California (US); Kolente virus from the Jones's roundleaf bat in Guinea;[60] Mount Elgon bat virus from the eloquent horseshoe bat in Kenya; Oita virus from the little Japanese horseshoe bat; and Fikirini virus from the striped leaf-nosed bat in Kenya.[61]
Orthomyxoviruses
[edit]
Orthomyxoviruses include influenza viruses. While birds are the primary reservoir for the genus Alphainfluenzavirus, a few bat species in Central and South America have also tested positive for the viruses. These species include the little yellow-shouldered bat and the flat-faced fruit-eating bat. Bat populations tested in Guatemala and Peru had high seropositivity rates, which suggests that influenza A infections are common among bats in the New World.[18]
Paramyxoviruses
[edit]Hendra, Nipah, and Menangle viruses
[edit]
Paramyxoviridae is a family that includes several zoonotic viruses naturally found in bats. Two are in the genus Henipavirus—Hendra virus and Nipah virus. Hendra virus was first identified in 1994 in Hendra, Australia. Four different species of flying fox have tested positive for Hendra virus: the gray-headed flying fox, little red flying fox, spectacled flying fox, and black flying fox.[62] Horses are the intermediate host between flying foxes and humans. Between 1994 and 2014, there were fifty-five outbreaks of Hendra virus in Australia, resulting in the death or euthanization of eighty-eight horses. Seven humans are known to have been infected by Hendra virus, with four fatalities.[16] Six of the seven infected humans were directly exposed to the blood or other fluids of sick or dead horses (three were veterinarians), while the seventh case was a veterinary nurse who had recently irrigated the nasal cavity of a horse not yet exhibiting symptoms. It is unclear how horses become infected with Hendra virus, though it is believed to occur following direct exposure to flying fox fluids. There is also evidence of horse-to-horse transmission. In late 2012, a vaccine was released to prevent infection in horses.[62] Vaccine uptake has been low, with an estimated 11–17% of Australian horses vaccinated by 2017.[63]
The first human outbreak of Nipah virus was in 1998 in Malaysia.[16] It was determined that flying foxes were also the reservoir of the virus, with domestic pigs as the intermediate host between bats and humans. Outbreaks have also occurred in Bangladesh, India, Singapore, and the Philippines. In Bangladesh, the primary mode of transmission of Nipah virus to humans is through the consumption of date palm sap. Pots set out to collect the sap are contaminated with flying fox urine and guano, and the bats also lick the sap streams flowing into the pots. It has been speculated that the virus may also be transmitted to humans by eating fruit partially consumed by flying foxes, or by coming into contact with their urine, though no definitive evidence supports this.[64]
An additional zoonotic paramyxovirus that bats harbor is Menangle virus, which was first identified at a hog farm in New South Wales, Australia. Flying foxes were once again identified as the natural reservoirs of the virus, with the black, spectacled, and gray-headed seropositive for the virus. Two employees of the hog farm became sick with flu-like illnesses, later shown to be a result of the virus.[16] Sosuga pararubulavirus is known to have infected one person—an American wildlife biologist who had conducted bat and rodent research in Uganda.[16] The Egyptian fruit bat later tested positive for the virus, indicating that it is potentially a natural reservoir.[65]
Other
[edit]Bats host several paramyxoviruses that are not known to affect humans. Bats are the reservoir of Cedar virus, a paramyxovirus first discovered in flying foxes South East Queensland.[16] The zoonotic potential of Cedar virus is unknown.[66] In Brazil in 1979, Mapuera orthorubulavirus was isolated from the saliva of the little yellow-shouldered bat. Mapuera virus has never been associated with disease in other animals or humans, but experimental exposure of mice to the virus resulted in fatality.[16] Tioman pararubulavirus has been isolated from the urine of the small flying fox, which causes fever in some domestic pigs after exposure, but no other symptoms. Tukoko virus has been detected from Leschenault's rousette in China.[16] Bats have been suggested as the host of Porcine orthorubulavirus, though definitive evidence has not been collected.[16]
Togaviruses
[edit]Togaviruses include alphaviruses, which have been detected in bats. Alphaviruses cause encephalitis in humans. Alphaviruses that have been detected in bats include Venezuelan equine encephalitis virus, Eastern equine encephalitis virus, and Western equine encephalitis virus. Sindbis virus has been detected from horseshoe bats and roundleaf bats. Chikungunya virus has been isolated from Leschenault's rousette, the Egyptian fruit bat, Sundevall's roundleaf bat, the little free-tailed bat, and Scotophilus species.[18]
Positive-sense single-stranded RNA viruses that replicate through a DNA intermediate
[edit]Retroviruses
[edit]Bats can be infected with retroviruses, including the gammaretrovirus found in horseshoe bats, Leschenault's rousette, and the greater false vampire bat. Several bat retroviruses have been identified that are similar to the Reticuloendotheliosis virus found in birds. These retroviruses were found in mouse-eared bats, horseshoe bats, and flying foxes. The discovery of varied and distinct gammaretroviruses in bat genomes indicates that bats likely played important roles in their diversification. Bats also host an extensive number of betaretroviruses, including within mouse-eared bats, horseshoe bats, and flying foxes. Bat betaretroviruses span the entire breadth of betaretrovirus diversity, similar to those of rodents, which may indicate that bats and rodents are primary reservoirs of the viruses. Betaretroviruses have infected bats for a majority of bat evolutionary history, since at least 36 million years ago.[67]
Double-stranded DNA viruses that replicate through a single-stranded RNA intermediate
[edit]
Hepadnaviruses
[edit]Hepadnaviruses are also known to affect bats, with the tent-making bat, Noack's roundleaf bat, and the halcyon horseshoe bat known to harbor several. The hepadnavirus found in the tent-making bat, which is a New World species, was the closest relative of human hepadnaviruses.[67] Though relatively few hepadnaviruses have been identified in bats, it is highly likely that additional strains will be discovered through further research. As of 2016, they had been found in four bat families: Hipposideridae and Rhinolophidae from the suborder Yinpterochiroptera and Molossidae and Vespertilionidae from Yangochiroptera. The high diversity of bat hosts suggests that bats share a long evolutionary history with hepadnaviruses, indicating bats may have had an important role in hepadnavirus evolution.[68]
See also
[edit]References
[edit]- ^ a b Calisher, C. H.; Childs, J. E.; Field, H. E.; Holmes, K. V.; Schountz, T. (2006). "Bats: Important Reservoir Hosts of Emerging Viruses". Clinical Microbiology Reviews. 19 (3): 531–545. doi:10.1128/CMR.00017-06. PMC 1539106. PMID 16847084.
- ^ Moratelli, Ricardo; Calisher, Charles H. (2015). "Bats and zoonotic viruses: Can we confidently link bats with emerging deadly viruses?". Memórias do Instituto Oswaldo Cruz. 110 (1): 1–22. doi:10.1590/0074-02760150048. PMC 4371215. PMID 25742261.
An increasingly asked question is 'can we confidently link bats with emerging viruses?'. No, or not yet, is the qualified answer based on the evidence available.
- ^ a b MacKenzie, John S.; Smith, David W. (2020). "COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don't". Microbiology Australia. 41: 45. doi:10.1071/MA20013. PMC 7086482. PMID 32226946.
Evidence from the sequence analyses clearly indicates that the reservoir host of the virus was a bat, probably a Chinese or Intermediate horseshoe bat, and it is probable that, like SARS-CoV, an intermediate host was the source of the outbreak.
- ^ a b Olival, Kevin J.; Weekley, Cristin C.; Daszak, Peter (2015). "Are Bats Really 'Special' as Viral Reservoirs? What We Know and Need to Know". Bats and Viruses. pp. 281–294. doi:10.1002/9781118818824.ch11. ISBN 978-1118818824.
- ^ a b Mollentze, Nardus; Streicker, Daniel G. (2020). "Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts". Proceedings of the National Academy of Sciences. 117 (17): 9423–9430. Bibcode:2020PNAS..117.9423M. doi:10.1073/pnas.1919176117. PMC 7196766. PMID 32284401.
- ^ a b c Letko, Michael; Seifert, Stephanie N.; Olival, Kevin J.; Plowright, Raina K.; Munster, Vincent J. (2020). "Bat-borne virus diversity, spillover and emergence". Nature Reviews Microbiology. 18 (8): 461–471. doi:10.1038/s41579-020-0394-z. PMC 7289071. PMID 32528128.
- ^ a b c d e f g h i Hayman, David T.S. (2016). "Bats as Viral Reservoirs". Annual Review of Virology. 3 (1): 77–99. doi:10.1146/annurev-virology-110615-042203. PMID 27578437.
- ^ a b c d e Xie, Jiazheng; Li, Yang; Shen, Xurui; Goh, Geraldine; Zhu, Yan; Cui, Jie; Wang, Lin-Fa; Shi, Zheng-Li; Zhou, Peng (2018). "Dampened STING-Dependent Interferon Activation in Bats". Cell Host & Microbe. 23 (3): 297–301.e4. doi:10.1016/j.chom.2018.01.006. PMC 7104992. PMID 29478775.
- ^ Gorman, James (28 January 2020). "How Do Bats Live With So Many Viruses?". The New York Times. Retrieved 17 March 2020.
- ^ Kuno, Goro (2001). "Persistence of arboviruses and antiviral antibodies in vertebrate hosts: its occurrence and impacts". Reviews in Medical Virology. 11 (3): 165–190. doi:10.1002/rmv.314. PMID 11376480. S2CID 22591717.
- ^ Sarkar, Saurav K.; Chakravarty, Ashim K. (1991). "Analysis of immunocompetent cells in the bat, Pteropus giganteus: Isolation and scanning electron microscopic characterization". Developmental & Comparative Immunology. 15 (4): 423–430. doi:10.1016/0145-305x(91)90034-v. PMID 1773865.
- ^ Barber, Glen N. (2015). "STING: Infection, inflammation and cancer". Nature Reviews Immunology. 15 (12): 760–770. doi:10.1038/nri3921. PMC 5004891. PMID 26603901.
- ^ Ahn, Matae; Anderson, Danielle E.; Zhang, Qian; Tan, Chee Wah; Lim, Beng Lee; Luko, Katarina; Wen, Ming; Chia, Wan Ni; Mani, Shailendra; Wang, Loo Chien; et al. (2019). "Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host". Nature Microbiology. 4 (5): 789–799. doi:10.1038/s41564-019-0371-3. PMC 7096966. PMID 30804542.
- ^ Yong, Kylie Su Mei; Ng, Justin Han Jia; Her, Zhisheng; Hey, Ying Ying; Tan, Sue Yee; Tan, Wilson Wei Sheng; Irac, Sergio Erdal; Liu, Min; Chan, Xue Ying; Gunawan, Merry; et al. (2018). "Bat-mouse bone marrow chimera: A novel animal model for dissecting the uniqueness of the bat immune system". Scientific Reports. 8 (1): 4726. Bibcode:2018NatSR...8.4726Y. doi:10.1038/s41598-018-22899-1. PMC 5856848. PMID 29549333.
- ^ a b c Joffrin, Léa; Dietrich, Muriel; Mavingui, Patrick; Lebarbenchon, Camille (2018). "Bat pathogens hit the road: But which one?". PLOS Pathogens. 14 (8): e1007134. doi:10.1371/journal.ppat.1007134. PMC 6085074. PMID 30092093.
- ^ a b c d e f g h i Anderson, Danielle E.; Marsh, Glenn A. (2015). "Bat Paramyxoviruses". Bats and Viruses. pp. 99–126. doi:10.1002/9781118818824.ch4. ISBN 978-1118818824.
- ^ Young, Cristin C. W.; Olival, Kevin J. (2016). "Optimizing Viral Discovery in Bats". PLOS ONE. 11 (2): e0149237. Bibcode:2016PLoSO..1149237Y. doi:10.1371/journal.pone.0149237. PMC 4750870. PMID 26867024.
- ^ a b c d e f g h i j k l m n Queen, Krista; Shi, Mang; Anderson, Larry J.; Tong, Suxiang (2015). "Other Bat-Borne Viruses". Bats and Viruses. pp. 217–247. doi:10.1002/9781118818824.ch9. ISBN 9781118818824.
- ^ "ICTV Master Species List 2018b.v2". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 30 March 2019. Retrieved 19 June 2019.
- ^ Cibulski, S. P.; Teixeira, T. F.; De Sales Lima, F. E.; Do Santos, H. F.; Franco, A. C.; Roehe, P. M. (2014). "A Novel Anelloviridae Species Detected in Tadarida brasiliensis Bats: First Sequence of a Chiropteran Anellovirus". Genome Announcements. 2 (5). doi:10.1128/genomeA.01028-14. PMC 4214982. PMID 25359906.
- ^ De Souza, William Marciel; Fumagalli, Marcílio Jorge; De Araujo, Jansen; Sabino-Santos, Gilberto; Maia, Felipe Gonçalves Motta; Romeiro, Marilia Farignoli; Modha, Sejal; Nardi, Marcello Schiavo; Queiroz, Luzia Helena; Durigon, Edison Luiz; et al. (2018). "Discovery of novel anelloviruses in small mammals expands the host range and diversity of the Anelloviridae". Virology. 514: 9–17. doi:10.1016/j.virol.2017.11.001. hdl:11449/165970. PMID 29128758.
- ^ a b Lecis, Roberta; Mucedda, Mauro; Pidinchedda, Ermanno; Zobba, Rosanna; Pittau, Marco; Alberti, Alberto (2020). "Genomic characterization of a novel bat-associated Circovirus detected in European Miniopterus schreibersii bats". Virus Genes. 56 (3): 325–328. doi:10.1007/s11262-020-01747-3. PMC 7088871. PMID 32088806.
- ^ Han, H.-J.; Wen, H.-L.; Zhao, L.; Liu, J.-W.; Luo, L.-M.; Zhou, C.-M.; Qin, X.-R.; Zhu, Y.-L.; Liu, M.-M.; Qi, R.; et al. (2017). "Novel coronaviruses, astroviruses, adenoviruses and circoviruses in insectivorous bats from northern China". Zoonoses and Public Health. 64 (8): 636–646. doi:10.1111/zph.12358. PMC 7165899. PMID 28371451.
- ^ Lorusso, Alessio; Teodori, Liana; Leone, Alessandra; Marcacci, Maurilia; Mangone, Iolanda; Orsini, Massimiliano; Capobianco-Dondona, Andrea; Camma’, Cesare; Monaco, Federica; Savini, Giovanni (2015). "A new member of the Pteropine Orthoreovirus species isolated from fruit bats imported to Italy". Infection, Genetics and Evolution. 30: 55–58. doi:10.1016/j.meegid.2014.12.006. PMID 25497353.
- ^ a b c d e Kohl, Claudia; Kurth, Andreas (2015). "Bat Reoviruses". Bats and Viruses. pp. 203–215. doi:10.1002/9781118818824.ch8. ISBN 9781118818824.
- ^ Tan, Yeh Fong; Teng, Cheong Lieng; Chua, Kaw Bing; Voon, Kenny (2017). "Pteropine orthoreovirus: An important emerging virus causing infectious disease in the tropics?". The Journal of Infection in Developing Countries. 11 (3): 215–219. doi:10.3855/jidc.9112. PMID 28368854.
- ^ Kocher, Jacob F.; Lindesmith, Lisa C.; Debbink, Kari; Beall, Anne; Mallory, Michael L.; Yount, Boyd L.; Graham, Rachel L.; Huynh, Jeremy; Gates, J. Edward; Donaldson, Eric F.; et al. (2018). "Bat Caliciviruses and Human Noroviruses Are Antigenically Similar and Have Overlapping Histo-Blood Group Antigen Binding Profiles". mBio. 9 (3). doi:10.1128/mBio.00869-18. PMC 5964351. PMID 29789360.
- ^ a b c d e f Ge, Xing-Yi; Hu, Ben; Shi, Zheng-Li (2015). "Bat Coronaviruses". Bats and Viruses. pp. 127–155. doi:10.1002/9781118818824.ch5. ISBN 978-1118818824.
- ^ Zhou, Peng; Yang, Xing-Lou; Wang, Xian-Guang; Hu, Ben; Zhang, Lei; Zhang, Wei; Si, Hao-Rui; Zhu, Yan; Li, Bei; Huang, Chao-Lin; et al. (2020). "A pneumonia outbreak associated with a new coronavirus of probable bat origin". Nature. 579 (7798): 270–273. Bibcode:2020Natur.579..270Z. doi:10.1038/s41586-020-2012-7. PMC 7095418. PMID 32015507.
- ^ "Novel Coronavirus (2019-nCoV) Situation Report – 22" (PDF). World Health Organization. 11 February 2020. Retrieved 15 February 2020.
- ^ Lu, Guangwen; Wang, Qihui; Gao, George F. (2015). "Bat-to-human: Spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond". Trends in Microbiology. 23 (8): 468–478. doi:10.1016/j.tim.2015.06.003. PMC 7125587. PMID 26206723.
- ^ "Middle East respiratory syndrome coronavirus (MERS-CoV)". World Health Organization. November 2019. Retrieved 5 April 2020.
- ^ "Middle East respiratory syndrome coronavirus (MERS-CoV)". World Health Organization. 11 March 2019. Retrieved 5 April 2020.
- ^ Nsikan, Akpan (21 January 2020). "New coronavirus can spread between humans – but it started in a wildlife market". National Geographic. Archived from the original on 22 January 2020. Retrieved 23 January 2020.
- ^ Fenton, M. Brock; Mubareka, Samira; Tsang, Susan M.; Simmons, Nancy B.; Becker, Daniel J. (2020). "COVID-19 and threats to bats". Facets. 5: 349–352. doi:10.1139/facets-2020-0028.
- ^ Aizenman, Nurith (20 February 2020). "New Research: Bats Harbor Hundreds Of Coronaviruses, And Spillovers Aren't Rare". NPR. Retrieved 5 April 2020.
- ^ Wacharapluesadee, Supaporn; Duengkae, Prateep; Rodpan, Apaporn; Kaewpom, Thongchai; Maneeorn, Patarapol; Kanchanasaka, Budsabong; Yingsakmongkon, Sangchai; Sittidetboripat, Nuntaporn; Chareesaen, Chaiyaporn; Khlangsap, Nathawat; et al. (2015). "Diversity of coronavirus in bats from Eastern Thailand". Virology Journal. 12: 57. doi:10.1186/s12985-015-0289-1. PMC 4416284. PMID 25884446.
- ^ a b c d e f Arai, Satoru; Aoki, Keita; Sơn, Nguyễn Trường; Tú, Vương Tân; Kikuchi, Fuka; Kinoshita, Gohta; Fukui, Dai; Thành, Hoàng Trung; Gu, Se Hun; Yoshikawa, Yasuhiro; et al. (2019). "Đakrông virus, a novel mobatvirus (Hantaviridae) harbored by the Stoliczka's Asian trident bat (Aselliscus stoliczkanus) in Vietnam". Scientific Reports. 9 (1): 10239. Bibcode:2019NatSR...910239A. doi:10.1038/s41598-019-46697-5. PMC 6629698. PMID 31308502.
- ^ Xu, Lin; Wu, Jianmin; He, Biao; Qin, Shaomin; Xia, Lele; Qin, Minchao; Li, Nan; Tu, Changchun (2015). "Novel hantavirus identified in black-bearded tomb bats, China". Infection, Genetics and Evolution. 31: 158–160. doi:10.1016/j.meegid.2015.01.018. PMC 7172206. PMID 25643870.
- ^ a b c d e f g Maganga, Gael Darren; Rougeron, Virginie; Leroy, Eric Maurice (2015). "Bat Filoviruses". Bats and Viruses. pp. 157–175. doi:10.1002/9781118818824.ch6. ISBN 9781118818824.
- ^ a b "Ebola virus disease". World Health Organization. 10 February 2020. Retrieved 13 April 2020.
- ^ a b "What is Ebola Virus Disease?". Centers for Disease Control and Prevention. 5 November 2019. Retrieved 13 April 2020.
Scientists do not know where Ebola virus comes from.
- ^ Rewar, Suresh; Mirdha, Dashrath (2015). "Transmission of Ebola Virus Disease: An Overview". Annals of Global Health. 80 (6): 444–451. doi:10.1016/j.aogh.2015.02.005. PMID 25960093.
Despite concerted investigative efforts, the natural reservoir of the virus is unknown.
- ^ a b Baseler, Laura; Chertow, Daniel S.; Johnson, Karl M.; Feldmann, Heinz; Morens, David M. (2017). "The Pathogenesis of Ebola Virus Disease". Annual Review of Pathology: Mechanisms of Disease. 12: 387–418. doi:10.1146/annurev-pathol-052016-100506. PMID 27959626.
The geographic ranges of many animal species, including bats, squirrels, mice and rats, dormice, and shrews, match or overlap with known outbreak sites of African filoviruses, but none of these mammals has yet been universally accepted as an EBOV reservoir.
- ^ von Csefalvay, Chris (2023), "Host-vector and multihost systems", Computational Modeling of Infectious Disease, Elsevier, pp. 121–149, doi:10.1016/b978-0-32-395389-4.00013-x, ISBN 978-0-323-95389-4, retrieved 2 March 2023
- ^ Olivero, Jesús; Fa, John E.; Real, Raimundo; Farfán, Miguel Ángel; Márquez, Ana Luz; Vargas, J. Mario; Gonzalez, J. Paul; Cunningham, Andrew A.; Nasi, Robert (2017). "Mammalian biogeography and the Ebola virus in Africa" (PDF). Mammal Review. 47: 24–37. doi:10.1111/mam.12074.
We found published evidence from cases of serological and/or polymerase chain reaction (PCR) positivity of EVD in non- human mammal, or of EVD-linked mortality, in 28 mammal species: 10 primates, three rodents, one shrew, eight bats, one carnivore, and five ungulates
- ^ a b c d "Marburg virus disease". World Health Organization. 15 February 2018. Retrieved 14 April 2020.
- ^ Yang, Xing-Lou; Tan, Chee Wah; Anderson, Danielle E.; Jiang, Ren-Di; Li, Bei; Zhang, Wei; Zhu, Yan; Lim, Xiao Fang; Zhou, Peng; Liu, Xiang-Ling; et al. (2019). "Characterization of a filovirus (Měnglà virus) from Rousettus bats in China". Nature Microbiology. 4 (3): 390–395. doi:10.1038/s41564-018-0328-y. PMID 30617348. S2CID 57574565.
- ^ a b c "Filoviruses – Ebola and Marburg Viruses". BU Research Support. 12 June 2019. Retrieved 14 April 2020.
- ^ Edwards, Megan R.; Basler, Christopher F. (2019). "Current status of small molecule drug development for Ebola virus and other filoviruses". Current Opinion in Virology. 35: 42–56. doi:10.1016/j.coviro.2019.03.001. PMC 6556423. PMID 31003196.
- ^ a b c d e f g h i j Kuzmin, Ivan V.; Rupprecht, Charles E. (2015). "Bat Lyssaviruses". Bats and Viruses. pp. 47–97. doi:10.1002/9781118818824.ch3. ISBN 978-1118818824.
- ^ a b c d Banyard, Ashley C.; Hayman, David; Johnson, Nicholas; McElhinney, Lorraine; Fooks, Anthony R. (2011). "Bats and Lyssaviruses". Research Advances in Rabies. Advances in Virus Research. Vol. 79. pp. 239–289. doi:10.1016/B978-0-12-387040-7.00012-3. ISBN 978-0123870407. PMID 21601050.
- ^ Calisher, Charles H. (2015). "Viruses in Bats". Bats and Viruses. pp. 23–45. doi:10.1002/9781118818824.ch2. ISBN 978-1118818824.
- ^ Klug, BJ; Turmelle, AS; Ellison, JA; Baerwald, EF; Barclay, RM (2010). "Rabies prevalence in migratory tree-bats in Alberta and the influence of roosting ecology and sampling method on reported prevalence of rabies in bats". Journal of Wildlife Diseases. 47 (1): 64–77. doi:10.7589/0090-3558-47.1.64. PMID 21269998.
- ^ "Rabies". www.who.int. Retrieved 8 July 2020.
- ^ a b Calderón, Alfonso; Guzmán, Camilo; Mattar, Salim; Rodríguez, Virginia; Acosta, Arles; Martínez, Caty (2019). "Frugivorous bats in the Colombian Caribbean region are reservoirs of the rabies virus". Annals of Clinical Microbiology and Antimicrobials. 18 (1): 11. doi:10.1186/s12941-019-0308-y. PMC 6423830. PMID 30890183.
- ^ Brock Fenton, M.; Streicker, Daniel G.; Racey, Paul A.; Tuttle, Merlin D.; Medellin, Rodrigo A.; Daley, Mark J.; Recuenco, Sergio; Bakker, Kevin M. (2020). "Knowledge gaps about rabies transmission from vampire bats to humans". Nature Ecology & Evolution. 4 (4): 517–518. doi:10.1038/s41559-020-1144-3. PMC 7896415. PMID 32203471. S2CID 212732288.
- ^ a b Messenger, Sharon L.; Smith, Jean S.; Rupprecht, Charles E. (2002). "Emerging Epidemiology of Bat-Associated Cryptic Cases of Rabies in Humans in the United States". Clinical Infectious Diseases. 35 (6): 738–747. doi:10.1086/342387. PMID 12203172.
- ^ Suu-Ire, Richard; Begeman, Lineke; Banyard, Ashley C.; Breed, Andrew C.; Drosten, Christian; Eggerbauer, Elisa; Freuling, Conrad M.; Gibson, Louise; Goharriz, Hooman; Horton, Daniel L.; et al. (2018). "Pathogenesis of bat rabies in a natural reservoir: Comparative susceptibility of the straw-colored fruit bat (Eidolon helvum) to three strains of Lagos bat virus". PLOS Neglected Tropical Diseases. 12 (3): e0006311. doi:10.1371/journal.pntd.0006311. PMC 5854431. PMID 29505617.
- ^ Blasdell, Kim R.; Widen, Steven G.; Wood, Thomas G.; Holmes, Edward C.; Vasilakis, Nikos; Tesh, Robert B.; Walker, Peter J.; Guzman, Hilda; Firth, Cadhla (2015). "Ledantevirus: A Proposed New Genus in the Rhabdoviridae has a Strong Ecological Association with Bats". The American Journal of Tropical Medicine and Hygiene. 92 (2): 405–410. doi:10.4269/ajtmh.14-0606. PMC 4347348. PMID 25487727.
- ^ Walker, Peter J.; Firth, Cadhla; Widen, Steven G.; Blasdell, Kim R.; Guzman, Hilda; Wood, Thomas G.; Paradkar, Prasad N.; Holmes, Edward C.; Tesh, Robert B.; Vasilakis, Nikos (2015). "Evolution of Genome Size and Complexity in the Rhabdoviridae". PLOS Pathogens. 11 (2): e1004664. doi:10.1371/journal.ppat.1004664. PMC 4334499. PMID 25679389.
- ^ a b Middleton, Deborah (2014). "Hendra Virus". Veterinary Clinics of North America: Equine Practice. 30 (3): 579–589. doi:10.1016/j.cveq.2014.08.004. PMC 4252762. PMID 25281398.
- ^ Manyweathers, J.; Field, H.; Longnecker, N.; Agho, K.; Smith, C.; Taylor, M. (2017). ""Why won't they just vaccinate?" Horse owner risk perception and uptake of the Hendra virus vaccine". BMC Veterinary Research. 13 (1): 103. doi:10.1186/s12917-017-1006-7. PMC 5390447. PMID 28407738.
- ^ Aditi; Shariff, M. (2019). "Nipah virus infection: A review". Epidemiology and Infection. 147: e95. doi:10.1017/S0950268819000086. PMC 6518547. PMID 30869046.
- ^ Amman, Brian R.; Albariño, Cesar G.; Bird, Brian H.; Nyakarahuka, Luke; Sealy, Tara K.; Balinandi, Stephen; Schuh, Amy J.; Campbell, Shelly M.; Ströher, Ute; Jones, Megan E. B.; et al. (2015). "A Recently Discovered Pathogenic Paramyxovirus, Sosuga Virus, is Present in Rousettus aegyptiacus Fruit Bats at Multiple Locations in Uganda". Journal of Wildlife Diseases. 51 (3): 774–779. doi:10.7589/2015-02-044. PMC 5022529. PMID 25919464.
- ^ Laing, Eric D.; Navaratnarajah, Chanakha K.; Cheliout Da Silva, Sofia; Petzing, Stephanie R.; Xu, Yan; Sterling, Spencer L.; Marsh, Glenn A.; Wang, Lin-Fa; Amaya, Moushimi; Nikolov, Dimitar B.; et al. (2019). "Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus". Proceedings of the National Academy of Sciences. 116 (41): 20707–20715. Bibcode:2019PNAS..11620707L. doi:10.1073/pnas.1911773116. PMC 6789926. PMID 31548390.
- ^ a b Tachedjian, Gilda; Hayward, Joshua A.; Cui, Jie (2015). "Bats and Reverse Transcribing RNA and DNA Viruses". Bats and Viruses. pp. 177–201. doi:10.1002/9781118818824.ch7. ISBN 9781118818824.
- ^ Rasche, Andrea; Souza, Breno Frederico de Carvalho Dominguez; Drexler, Jan Felix (2016). "Bat hepadnaviruses and the origins of primate hepatitis B viruses". Current Opinion in Virology. 16: 86–94. doi:10.1016/j.coviro.2016.01.015. PMID 26897577.