Coenzyme B

 | |
| Names | |
|---|---|
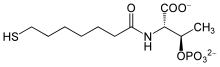
| IUPAC name 2-[(7-mercapto-1-oxoheptyl)amino]-3-phosphonooxybutanoic acid | |
| Identifiers | |
3D model (JSmol) | |
| ChemSpider | |
PubChem CID | |
| UNII | |
CompTox Dashboard (EPA) | |
| |
| |
| Properties | |
| C 11H 22NO 7PS | |
| Molar mass | 343.333641 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Coenzyme B is a coenzyme required for redox reactions in methanogens. The full chemical name of coenzyme B is 7-mercaptoheptanoylthreoninephosphate.[1] The molecule contains a thiol, which is its principal site of reaction.
Coenzyme B reacts with 2-methylthioethanesulfonate (methyl-Coenzyme M, abbreviated CH
3–S–CoM), to release methane in methanogenesis:[2]
- CH
3–S–CoM + HS–CoB → CH
4 + CoB–S–S–CoM
This conversion is catalyzed by the enzyme methyl coenzyme M reductase, which contains cofactor F430 as the prosthetic group.
A related conversion that utilizes both HS-CoB and HS-CoM is the reduction of fumarate to succinate, catalyzed by fumarate reductase:[3]
- HS–CoM + HS–CoB + −O
2CCH=CHCO−
2 → −O
2CCH
2–CH
2CO−
2 + CoB–S–S–CoM
Importance of Coenzyme B in Methanogenesis
[edit]Coenzyme B is an important component in the terminal step of methane biogenesis.[4] It acts as a two electron-donor to reduce coenzyme M (methyl-coenzyme) into two molecules a methane and a heterodisulfide.[5] Two separate experiment that were performed, one with coenzyme B and other without coenzyme B, indicated that using coenzyme B before the formation of the methane molecule, results in a more efficient and consistent bond cleavage.[6]
References
[edit]- ^ Noll KM, Rinehart KL, Tanner RS, Wolfe RS (1986). "Structure of component B (7-mercaptoheptanoylthreonine phosphate) of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum". Proceedings of the National Academy of Sciences. 83 (12): 4238–42. Bibcode:1986PNAS...83.4238N. doi:10.1073/pnas.83.12.4238. PMC 323707. PMID 3086878.
- ^ Thauer RK (September 1998). "Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture". Microbiology. 144 (Pt 9): 2377–406. doi:10.1099/00221287-144-9-2377. PMID 9782487.
- ^ Heim S, Künkel A, Thauer RK, Hedderich R (April 1998). "Thiol:fumarate reductase (Tfr) from Methanobacterium thermoautotrophicum—identification of the catalytic sites for fumarate reduction and thiol oxidation". European Journal of Biochemistry. 253 (1): 292–9. doi:10.1046/j.1432-1327.1998.2530292.x. PMID 9578488.
- ^ Dey, Mishtu; Li, Xianghui; Kunz, Ryan C; Ragsdale, Stephen W (2010-12-22). "Detection of Organometallic and Radical Intermediates in the Catalytic Mechanism of Methyl-Coenzyme M Reductase Using the Natural Substrate Methyl-Coenzyme M and a Coenzyme B Substrate Analogue". Biochemistry. 49 (51): 10902–10911. doi:10.1021/bi101562m. PMID 21090696.[permanent dead link]
- ^ Cedervall, Peder E; Dey, Mishtu; Pearson, Arwen R; Ragsdale, Stephen W; Wilmot, Carrie M (2010-07-22). "Structural Insight into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues". Biochemistry. 49 (35): 7683–7693. doi:10.1021/bi100458d. PMC 3098740. PMID 20707311.
- ^ Horng, Yih-Chern; Becker, Donald F; Ragsdale, Stephen W (2001-10-30). "Mechanistic Studies of Methane Biogenesis by Methyl-Coenzyme M Reductase: Evidence that Coenzyme B Participates in Cleaving the C−S Bond of Methyl-Coenzyme M". Biochemistry. 40 (43): 12875–12885. doi:10.1021/bi011196y. PMID 11669624.[permanent dead link]