KIX domain

| KIX (CBP) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
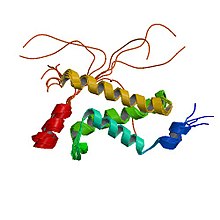 Complex between the KIX domain of CBP (yellow, green and cyan) and the C-terminal transactivation domain of p65/RELA (red)[1] | |||||||||
| Identifiers | |||||||||
| Symbol | KIX | ||||||||
| Pfam | PF02172 | ||||||||
| Pfam clan | CL0589 | ||||||||
| InterPro | IPR003101 | ||||||||
| CATH | 1sb0 | ||||||||
| SCOP2 | 1sb0 / SCOPe / SUPFAM | ||||||||
| |||||||||
| KIX (MED15) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | KIX_2 | ||||||||
| Pfam | PF16987 | ||||||||
| InterPro | IPR036546 | ||||||||
| |||||||||
In biochemistry, the KIX domain (kinase-inducible domain (KID) interacting domain) or CREB binding domain is a protein domain of the eukaryotic transcriptional coactivators CBP and P300. It serves as a docking site for the formation of heterodimers between the coactivator and specific transcription factors. Structurally, the KIX domain is a globular domain consisting of three α-helices and two short 310-helices.
The KIX domain was originally discovered in 1996 as the specific and minimal region in CBP that binds and interacts with phosphorylated CREB to activate transcription.[2] It was thus first termed CREB-binding domain. However, when it was later discovered that it also binds many other proteins, the more general name KIX domain became favoured. The KIX domain contains two separate binding sites: the "c-Myb site", named after the oncoprotein c-Myb, and the "MLL site", named after the proto-oncogene MLL (Mixed Lineage Leukemia, KMT2A).[3]
The paralogous coactivators CBP (CREBBP) and P300 (EP300) are recruited to DNA-bound transcription factors to activate transcription. Coactivators can associate with promoters and enhancers in the DNA only indirectly through protein-protein contacts with transcription factors. CBP and P300 activate transcription synergistically in two ways: first, by remodelling and relaxing chromatin through their intrinsic histone acetyltransferase activity, and second, by recruiting the basal transcription machinery, such as RNA polymerase II.[4]
The KIX domain belongs to the proposed GACKIX domain superfamily. GACKIX comprises structurally and functionally highly homologous domains in related proteins. It is named after the protein GAL11 / ARC105 (MED15), the plant protein CBP-like, and the KIX domain from CBP and P300.[5] Additional instances include RECQL5 and related plant proteins.[6][7] All of these contain a KIX domain or KIX-related domain that interacts with the transactivation domain of many different transcription factors. The distinction between a KIX domain, a KIX-related domain and a GACKIX domain is subject to an ongoing debate and not clearly defined.
The full CBP/P300 protein
[edit]
Aside from the KIX domain, CBP and P300 contain many other protein binding domains that should not be confused (numbers are aa numberings):
- CH1/TAZ1 domain, CBP[347–433], P300[323-423][8]
- KIX domain, CBP[587–666], P300[566–645][8]
- Bromodomain, CBP[1103–1175], P300[1067–1139][8]
- CH2 domain (IPR010303), CBP[1191–1317], P300[1155-1280].[9]
- HAT domain, CBP[1323–1700], P300[1287–1663][8]
- CH3/ZZ domain, CBP[1701-1744], P300[1664-1707][8]
- CH3/TAZ2 domain, CBP[1765–1846], P300[1728-1809][8]
- IRF-3 binding (i-BiD),[10] nuclear receptor coactivator binding (NCBD),[11] or SRC1 interaction domain (SID; IPR014744),[12] CBP[2020-2113], P300[1992-2098].
All three CH (cysteine/histidine-rich) domains are zinc fingers.[9]
Interactions
[edit]
Human and animal proteins:
- ARNTL (BMAL1)[13][14]
- ATF1[15]
- ATF4 (CREB2)[15]
- BRCA1[4][15]
- CREB[10]
- Cubitus interruptus (in D. melanogaster)[4][15]
- ELK4 (SAP1)[15]
- FOXO3 (FOXO3a)[16]
- GLI3[15][17]
- JUN (c-Jun)[4][15]
- KMT2A (MLL)[18][4]
- MYB (c-Myb)[4][15]
- NFE2 (NF-E2 p45)[15]
- RELA (NF-κB p65)[1][19]
- SREBP[4][15]
- STAT1[4]
- TCF3 (E2A)[20]
- TP53 (p53)[4]
- YY1[15]
Yeast proteins:
Viral proteins:
- Regulatory protein E2 (from Human papillomavirus)[4][15]
- Tat (from HIV-1)[4]
- Tax (from Deltaretrovirus)[4][15]
References
[edit]- ^ a b Lecoq L, Raiola L, Chabot PR, Cyr N, Arseneault G, Legault P, Omichinski JG (May 2017). "Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-κB and transcription regulatory factors". Nucleic Acids Research. 45 (9): 5564–5576. doi:10.1093/nar/gkx146. PMC 5435986. PMID 28334776.
- ^ Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, et al. (February 1996). "Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism". Molecular and Cellular Biology. 16 (2): 694–703. doi:10.1128/MCB.16.2.694. PMC 231049. PMID 8552098.
- ^ Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE (November 2002). "Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain". The Journal of Biological Chemistry. 277 (45): 43168–74. CiteSeerX 10.1.1.615.9401. doi:10.1074/jbc.M207660200. PMID 12205094.
- ^ a b c d e f g h i j k l m n o p q Thakur JK, Yadav A, Yadav G (February 2014). "Molecular recognition by the KIX domain and its role in gene regulation". Nucleic Acids Research. 42 (4): 2112–25. doi:10.1093/nar/gkt1147. PMC 3936767. PMID 24253305.
- ^ Novatchkova M, Eisenhaber F (January 2004). "Linking transcriptional mediators via the GACKIX domain super family". Current Biology. 14 (2): R54-5. Bibcode:2004CBio...14..R54N. doi:10.1016/j.cub.2003.12.042. PMID 14738747.
- ^ Popuri V, Ramamoorthy M, Tadokoro T, Singh DK, Karmakar P, Croteau DL, Bohr VA (July 2012). "Recruitment and retention dynamics of RECQL5 at DNA double strand break sites". DNA Repair. 11 (7): 624–35. doi:10.1016/j.dnarep.2012.05.001. PMC 3374033. PMID 22633600.
- ^ Yadav A, Thakur JK, Yadav G (November 2017). "KIXBASE: A comprehensive web resource for identification and exploration of KIX domains". Scientific Reports. 7 (1): 14924. Bibcode:2017NatSR...714924Y. doi:10.1038/s41598-017-14617-0. PMC 5668377. PMID 29097748.
- ^ a b c d e f UniProt and InterPro entry for CBP (Q92793) and P300 (Q09472)
- ^ a b Park S, Martinez-Yamout MA, Dyson HJ, Wright PE (August 2013). "The CH2 domain of CBP/p300 is a novel zinc finger". FEBS Letters. 587 (16): 2506–11. Bibcode:2013FEBSL.587.2506P. doi:10.1016/j.febslet.2013.06.051. PMC 3765250. PMID 23831576.
- ^ a b Lin CH, Hare BJ, Wagner G, Harrison SC, Maniatis T, Fraenkel E (September 2001). "A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies". Molecular Cell. 8 (3): 581–90. doi:10.1016/S1097-2765(01)00333-1. PMID 11583620.
- ^ Demarest SJ, Deechongkit S, Dyson HJ, Evans RM, Wright PE (January 2004). "Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein". Protein Science. 13 (1): 203–10. doi:10.1110/ps.03366504. PMC 2286511. PMID 14691235.
- ^ Sheppard HM, Harries JC, Hussain S, Bevan C, Heery DM (January 2001). "Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1". Molecular and Cellular Biology. 21 (1): 39–50. doi:10.1128/MCB.21.1.39-50.2001. PMC 86566. PMID 11113179.
- ^ Takahata S, Ozaki T, Mimura J, Kikuchi Y, Sogawa K, Fujii-Kuriyama Y (September 2000). "Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3". Genes to Cells. 5 (9): 739–47. doi:10.1046/j.1365-2443.2000.00363.x. PMID 10971655. S2CID 41625860.
- ^ Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, et al. (June 2015). "Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus". Nature Structural & Molecular Biology. 22 (6): 476–484. doi:10.1038/nsmb.3018. PMC 4456216. PMID 25961797.
- ^ a b c d e f g h i j k l m Vo N, Goodman RH (April 2001). "CREB-binding protein and p300 in transcriptional regulation". The Journal of Biological Chemistry. 276 (17): 13505–8. doi:10.1074/jbc.R000025200. PMID 11279224.
- ^ Wang F, Marshall CB, Li GY, Yamamoto K, Mak TW, Ikura M (December 2009). "Synergistic interplay between promoter recognition and CBP/p300 coactivator recruitment by FOXO3a". ACS Chemical Biology. 4 (12): 1017–27. doi:10.1021/cb900190u. PMID 19821614.
- ^ Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S (March 1999). "Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3". The Journal of Biological Chemistry. 274 (12): 8143–52. doi:10.1074/jbc.274.12.8143. PMID 10075717.
- ^ Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ (April 2001). "MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein". Molecular and Cellular Biology. 21 (7): 2249–58. doi:10.1128/MCB.21.7.2249-2258.2001. PMC 86859. PMID 11259575.
- ^ Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T (April 1997). "CREB-binding protein/p300 are transcriptional coactivators of p65". Proceedings of the National Academy of Sciences of the United States of America. 94 (7): 2927–32. Bibcode:1997PNAS...94.2927G. doi:10.1073/pnas.94.7.2927. PMC 20299. PMID 9096323.
- ^ Bradney C, Hjelmeland M, Komatsu Y, Yoshida M, Yao TP, Zhuang Y (January 2003). "Regulation of E2A activities by histone acetyltransferases in B lymphocyte development". The Journal of Biological Chemistry. 278 (4): 2370–6. doi:10.1074/jbc.M211464200. PMID 12435739.
External links
[edit]- KIXBASE, a database of KIX-superfamily domains and PDB structures (HMM training dataset)