C11orf49
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
| CSTPP1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | CSTPP1, chromosome 11 open reading frame 49, C11orf49, centriolar satellite-associated tubulin polyglutamylase complex regulator 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1915079 HomoloGene: 11471 GeneCards: CSTPP1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
C11orf49 is a protein coding gene that in humans encodes for the C11orf49 protein.[5] It is heavily expressed in brain tissue and peripheral blood mononuclear cells, with the latter being an important component of the immune system.[6][7] It is predicted that the C11orf49 protein acts as a kinase, and has been shown to interact with HTT (determining protein for Huntington's disease) and APOE2 (risk protein for Alzheimer's).[8][9]
Gene[edit]
Aliases[edit]
Common aliases are UPF0705, FLJ22210, and MGC4707.[7]
Location[edit]
C11orf49 is found at locus p11.2 on human chromosome 11, with a plus strand orientation.[7] The gene is 224,830 bp long including introns, and spans from position 46,936,806 to 47,161,635 on chromosome 11. [10]
Transcript Variants[edit]
There are 7 known transcript variants for the mRNA of C11orf49, with variant 2 encoding for the most complete protein. Variant 1 lacks a 3’ splice junction, which results in a truncated 3’ terminus compared to variant 2. Variant 3 contains an alternate splice site at the 3’ end, which lacks an internal region near the 3’ terminus compared to variant 2. Variant 4 has an alternate 3’ terminus exon, resulting in a truncated 3’ terminus compared to variant 2. Variant 5 lacks an exon in the 5’ coding region which results in an upstream start codon, and has alternate splice site near the 3’ region. This results in a distinct N-terminus and a missing internal region near the 3’ terminus compared to variant 2. Variants 6 and 7 are both represented as candidates for nonsense-mediated mRNA decay (NMD), and do not encode for viable proteins.[5]
| Name | Accession Number | Numbers of Exons | Size (bp) |
| Transcript Variant 1 | NM_001003676.3 | 8 | 1923 |
| Transcript Variant 2 | NM_001003677.3 | 9 | 1668 |
| Transcript Variant 3 | NM_024113.5 | 8 | 1650 |
| Transcript Variant 4 | NM_001003678.3 | 9 | 1159 |
| Transcript Variant 5 | NM_001278222.1 | 8 | 1619 |
| Transcript Variant 6 | NR_103471.2 | 10 | 1895 |
| Transcript Variant 7 | NR_103472.2 | 8 | 1519 |
Table 1. Known human mRNA transcript variants for C11orf49.
Protein[edit]



Isoforms[edit]
There are 5 known isoforms for the C11orf49 protein with isoform 2 being the most complete protein, encoded by transcript variant 2.[5]
| Name | Accession Number | Size (AA) |
| Isoform 1 | NP_001003676.1 | 274 |
| Isoform 2 | NP_001003677.1 | 337 |
| Isoform 3 | NP_077018.1 | 331 |
| Isoform 4 | NP_001003678.1 | 326 |
| Isoform 5 | NP_001265151.1 | 322 |
Table 2. Known human protein isoforms for C11orf49.
Composition[edit]
The C11orf49 protein has a molecular weight of 38.1 kD, and an isoelectric point of about pH = 5.[11] Protein composition falls under normal levels for each amino acid, and there are no conserved repeats, patterns, or charged clusters to be seen. There are no hydrophobic or transmembrane regions to be seen.[12]
Protein Domain[edit]
The C11orf49 protein is predicted to contain a protein kinase domain near the N' terminus (residues 12-51)[8]
Secondary Structure[edit]
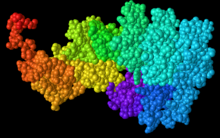
Secondary structure prediction tools such as Ali2D, Phyre2, and i-Tasser all predict that the C11orf49 protein is mostly composed of alpha helices, with no predicted beta sheets.[8][13][14] Information on where these alpha helices are located can be seen to the right of the page.
Tertiary Structure[edit]
i-Tasser predicted tertiary structure is included to the right of the page.[14]
Post-Translational Modifications[edit]
Phosphorylation[edit]
The C11orf49 protein is predicted to be phosphorylated at 4 different sites, mainly on serine residues, but also on one threonine residue.[15]
| Position | AA | Kinase |
| 310 | Serine | AGC/Akt |
| 48 | Threonine | AGC/Akt/AKT1 |
| 66 | Serine | AGC/Akt |
| 318 | Serine | AGC/Akt |
Table 3. Predicted phosphorylation sites for the C11orf49 human protein.
Sumoylation[edit]
The C11orf49 protein is predicted to be sumoylated at positions 119 and 320, both lysine residues.[15]
Subcellular Localization[edit]
The C11orf49 protein found in humans is predicted to be localized in the cytoplasm.[16]
Gene Level Regulation[edit]
Promoters[edit]

There are 7 promoters listed on Genomatix, however only one of the promoters (GXP_204543) starts at the beginning of the C11orf49 gene that is found in humans, and also has the greatest number of encoding transcripts.[17]
| Promoter ID | Start Position | End Position | Size (bp) | Orientation | Total # of transcripts |
| GXP_204543 | 46935524 | 46936819 | 1296 | plus strand | 32 |
| GXP_3162280 | 47050923 | 47051962 | 1040 | plus strand | 1 |
| GXP_3162281 | 47051454 | 47052500 | 1047 | plus strand | 2 |
| GXP_3162283 | 47136696 | 47137735 | 1040 | plus strand | 1 |
| GXP_3162284 | 47153395 | 47154434 | 1040 | plus strand | 1 |
| GXP_3162285 | 47153944 | 47154983 | 1040 | plus strand | 1 |
| GXP_204542 | 47159105 | 47160144 | 1040 | plus strand | 1 |
Table 4. List of promoters associated with the C11orf49 human gene.
Transcription Factors[edit]

The following transcription factors are predicted to bind to the GXP_204543 promoter. [18] The higher the matrix score, the more likely the transcription factor is to bind to the promoter. Information on where these transcription factors bind on the GXP_204543 promoter is showcased in the image to the right of the page.
| Matrix Family | Detailed Family Info | Detailed Matrix Info | Matrix Score |
| V$NKXH | NKX homeodomain factors | Homeodomain factor NKX-2.5 | 1 |
| V$GATA | GATA binding factor | GATA-binding factor 3 | 0.992 |
| V$LEFF | LEF1/TCF | Involved in the Wnt signal pathway | 0.991 |
| O$VTBP | Vertebrate TATA binding factor | Cellular and viral TATA box elements | 0.99 |
| V$KLFS | Krueppel like TFs | Gut-enriched Krueppel-like TF | 0.982 |
| V$MYBL | Cellular and Viral myb-like TFs | V-Myb | 0.978 |
| V$E2FF | E2F-myc activator | E2F TF 1 | 0.976 |
| V$MEF3 | MEF3 binding sites | Sine oculis homeobox homolog 2 | 0.972 |
| V$XBBF | X-box binding factors | X-box binding protein RFX1 | 0.966 |
| V$ETSF | Human and murine ETS1 factors | Elk-1 | 0.958 |
| V$PBXC | PBX-MEIS complexes | Pre-B-cell leukemia homeobox 3 | 0.949 |
| V$CAAT | CCAAT binding factors | Cellular and viral CCAAT box | 0.927 |
| V$HEAT | Heat shock factors | Heat shock factor 1 | 0.927 |
| V$MYT1 | MYT1 C2HC zinc finger protein | Myelin TF 1-like, neuronal C2H2 ZF 1 | 0.925 |
| V$GCMF | Chorion-specific TFs | Glial cells missing homolog 1 | 0.902 |
| V$ZF04 | C2H2 zinc finger TF 4 | Zinc finger and BTB domain | 0.9 |
| V$MAZF | Myc associated zinc fingers (MAZ) | MAZ | 0.875 |
| V$PAX9 | Pax-9 binding sites | Zebrafish Pax-9 binding site | 0.848 |
| V$DMRT | DM domain-containing TFs | Mab-3 related TF 1 | 0.817 |
Table 5. List of binding transcription factors to the GXP_204543 promoter.
Gene Expression[edit]


Tissue Specific Expression[edit]
Both microarray expression patterns and RNA-Seq data show very high levels of expression in the brain.[5][19] RNA-Seq data also shows high expression in lung fetal tissue.[5] Additional information for other tissues is included to the right of the page.
Conditions of Differentiated Expression[edit]
C11orf49 expression is significantly increased after the overexpression of claudin-1 in lung adenocarcinoma cell lines.[20] Claudin-1 specifically prevents paracellular diffusion of small molecules through tight junctions in the epidermis.
C11orf49 expression is significantly decreased after the treatment of camptothecin on a renal epithelial cell line.[21] Camptothecin is an alkaloid that inhibits the nuclear enzyme DNA topoisomerase, and has exhibited antitumor activity. It has also shown the ability to cause apoptosis by changing the permeability of the mitochondrial membrane, releasing cytochrome C.
Post-Transcription Regulation[edit]

5' UTR[edit]
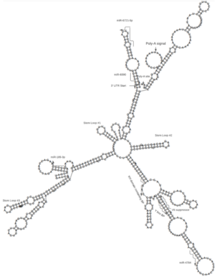
There is a predicted stem-loop structure in the 5' UTR of the C11orf49 transcript from nucleotides 15-26 shown to the right of the page.[22]
3' UTR[edit]
There are predicted stem-loop structures and miRNA binding sites for the 3' UTR of the C11orf49 transcript shown to the right of the page.[22][23]
Protein-Protein Interactions[edit]
The database provided by PSICQUIC indicates that the C11orf49 protein found in humans interacts with the following proteins listed in Table 6.[9] All interactions were determined using two-hybrid screening experiments.[9]
| Protein | Description |
| HTT | Huntingtin protein |
| APOE | Apolipoprotein E |
| PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit |
| FH | Fumarate hydratase |
| GCA | Grancalcin |
| PHF1 | PHD finger protein 1 |
| VPS54 | Vacuolar protein sorting-associated protein 54 |
| ZFHX3 | Zinc finger homeobox protein 3 |
| RAB7L1 | RAS oncogene family-like 1 |
| NDRG1 | Stress responsive protein |
| PNMA5 | Paraneoplastic antigen-like protein 5 |
| TXN2 | Thioredoxin |
Table 6. List of proteins that interact with the C11orf49 protein found in humans.
Homology and Evolution[edit]
Orthologs and Paralogs[edit]
C11orf49 can be found among a wide variety of taxonomic groups, including but not limited to Mammalia, Aves, Reptilia, Amphibia, Cyprinidae, Hemichordata, Cnidaria, Platyhelminthes, Arthropoda, Placozoa, Choanoflagellate, Spizellomyces, and Oomycota.[24][25] However, C11orf49 could not be found in Insecta or Plantae.[24][25] There are no known paralogs of C11orf49.[24][25]
| Genus and Species | Common name | Taxonomic group | Divergence (MYA) | Accession # | AA length | Identity (%) | Similarity (%) |
| Mus musculus | Mouse | Rodentia | 89 | NP_780332.1 | 331 | 92 | 95.5 |
| Gallus gallus | Chicken | Aves | 318 | XP_015142672.1 | 331 | 76.7 | 83.5 |
| Chelonia mydas | Green Sea Turtle | Reptilia | 318 | XP_007054360.2 | 362 | 73.4 | 79.9 |
| Geotrypetes seraphini | Gaboon caecilian | Amphibia | 352 | XP_033784118.1 | 329 | 69.9 | 81.7 |
| Xenopus tropicalis | Tropical Clawed Frog | Amphibia | 352 | NM_001079316.1 | 330 | 61.9 | 77.3 |
| Danio rerio | Zebrafish | Cyprinoidae | 433 | NP_001002479.1 | 331 | 54.7 | 72.2 |
| Sacoglossus kowalevskii | Acorn Worm | Hemichordata | 627 | XP_006821066.1 | 299 | 43.6 | 58.2 |
| Nematostella vectensis | Starlet Sea Anemone | Cnidaria | 687 | XP_032240720.1 | 300 | 42.4 | 57.3 |
| Macrostomum lignano | Flatworm | Platyhelminthes | 692 | PAA77967.1 | 404 | 29.7 | 42.3 |
| Stegodyphus mimosarum | African Velvet Spider | Arthropoda | 736 | KFM74201.1 | 321 | 24.3 | 38.5 |
| Trichoplax adhaerens | Trichoplax | Placozoa | 747 | XP_002108042.1 | 263 | 24.7 | 39.6 |
| Salpingoeca rosetta | N/A | Choanoflagellate | 928 | XP_004994083.1 | 231 | 15.5 | 25.5 |
| Spizellomyces palustris | Australian fungus | Spizellomyces | 1017 | TPX67906.1 | 298 | 21.4 | 32.7 |
| Saprolegnia diclina | Cotton Mould | Oomycota | 1552 | XP_008621502.1 | 314 | 22.9 | 35.6 |
Table 7. List of selected orthologs of C11orf49.
Evolution[edit]

History[edit]
Saprolegnia diclina is the most distantly related ortholog of C11orf49 known, with its divergence from ancestral humans approximately 1,552 MYA.[24][26]
Evolutionary Rate[edit]
After performing a molecular clock analysis, C11orf49 has evolved at a faster rate than Cytochrome c but slower than Fibrinogen alpha. The graph containing this analysis is to the right of the page.
Function[edit]
Protein Kinase Activity[edit]
C11orf49 is predicted to act as a cAMP-dependent protein kinase.[8]
Clinical Significance[edit]
C11orf49 has been shown to interact with proteins HTT and APOE2, which are associated with Huntington's disease and Alzheimer's, respectively.[9] Due to the predicted function of C11orf49, this interaction could be kinase-oriented.
C11orf49 expression is significantly increased after the overexpression of Claudin-1 in lung adenocarcinoma cells.[20]
C11orf49 expression is significantly decreased after the treatment of camptothecin on a renal epithelial cell line.[21]
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000149179 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000040591 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d e "C11orf49 chromosome 11 open reading frame 49 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-12-17.
- ^ "GDS596 / 203257_s_at". www.ncbi.nlm.nih.gov. Retrieved 2020-12-17.
- ^ a b c "C11orf49 Gene - GeneCards | CK049 Protein | CK049 Antibody". www.genecards.org. Retrieved 2020-12-17.
- ^ a b c d "Phyre 2 Results for Undefined". www.sbg.bio.ic.ac.uk. Retrieved 2020-12-17.[permanent dead link]
- ^ a b c d "PSICQUIC View". www.ebi.ac.uk. Retrieved 2020-12-17.
- ^ "Human BLAT Search". genome.ucsc.edu. Retrieved 2020-12-17.
- ^ "ExPASy - Compute pI/Mw tool". web.expasy.org. Retrieved 2020-12-17.
- ^ "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2020-12-18.
- ^ "Bioinformatics Toolkit". toolkit.tuebingen.mpg.de. Retrieved 2020-12-18.
- ^ a b "I-TASSER results". zhanglab.ccmb.med.umich.edu. Retrieved 2020-12-18.[permanent dead link]
- ^ a b "SIB Swiss Institute of Bioinformatics | Expasy". www.expasy.org. Retrieved 2020-12-18.
- ^ "PSORT II Prediction". psort.hgc.jp. Retrieved 2020-12-18.
- ^ "Genomatix: Retrieve and analyze promoters: Query Input". www.genomatix.de. Retrieved 2020-12-18.[permanent dead link]
- ^ "Genomatix: MatInspector Input". www.genomatix.de. Retrieved 2020-12-18.[permanent dead link]
- ^ "GDS596 / 203257_s_at". www.ncbi.nlm.nih.gov. Retrieved 2020-12-18.
- ^ a b "59175105 - GEO Profiles - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-12-19.
- ^ a b "14476184 - GEO Profiles - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-12-19.
- ^ a b "RNAfold web server". rna.tbi.univie.ac.at. Retrieved 2020-12-19.
- ^ "TargetScanHuman 7.2 predicted targeting of Human C11orf49". www.targetscan.org. Retrieved 2020-12-19.
- ^ a b c d "BLAST: Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2020-12-19.
- ^ a b c "Human BLAT Search". genome.ucsc.edu. Retrieved 2020-12-19.
- ^ "TimeTree :: The Timescale of Life". www.timetree.org. Retrieved 2020-12-19.




