Inner mitochondrial membrane
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
| Cell biology | |
|---|---|
| mitochondrion | |
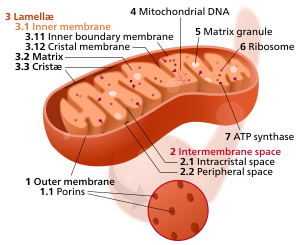 Components of a typical mitochondrion
3 Lamella
4 Mitochondrial DNA |
The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.
Structure
[edit]The structure of the inner mitochondrial membrane is extensively folded and compartmentalized. The numerous invaginations of the membrane are called cristae, separated by crista junctions from the inner boundary membrane juxtaposed to the outer membrane. Cristae significantly increase the total membrane surface area compared to a smooth inner membrane and thereby the available working space for oxidative phosphorylation.
The inner membrane creates two compartments. The region between the inner and outer membrane, called the intermembrane space, is largely continuous with the cytosol, while the more sequestered space inside the inner membrane is called the matrix.
Cristae
[edit]For typical liver mitochondria, the area of the inner membrane is about 5 times as large as the outer membrane due to cristae. This ratio is variable and mitochondria from cells that have a greater demand for ATP, such as muscle cells, contain even more cristae. Cristae membranes are studded on the matrix side with small round protein complexes known as F1 particles, the site of proton-gradient driven ATP synthesis. Cristae affect overall chemiosmotic function of mitochondria.[1]
Cristae junctions
[edit]Cristae and the inner boundary membranes are separated by junctions. The end of cristae are partially closed by transmembrane protein complexes that bind head to head and link opposing crista membranes in a bottleneck-like fashion.[2] For example, deletion of the junction protein IMMT leads to a reduced inner membrane potential and impaired growth[3] and to dramatically aberrant inner membrane structures which form concentric stacks instead of the typical invaginations.[4]
Composition
[edit]The inner membrane of mitochondria is similar in lipid composition to the membrane of bacteria. This phenomenon can be explained by the endosymbiont hypothesis of the origin of mitochondria as prokaryotes internalized by a eukaryotic host cell.
In pig heart mitochondria, phosphatidylethanolamine makes up the majority of the inner mitochondrial membrane at 37.0% of the phospholipid composition. Phosphatidylcholine makes up about 26.5%, cardiolipin 25.4%, and phosphatidylinositol 4.5%.[5] In S. cerevisiae mitochondria, phosphatidylcholine makes up 38.4% of the IMM, phosphatidylethanolamine makes up 24.0%, phosphatidylinositol 16.2%, cardiolipin 16.1%, phosphatidylserine 3.8%, and phosphatidic acid 1.5%.[6]
In the inner mitochondrial membrane, the protein-to-lipid ratio is 80:20, in contrast to the outer membrane, which is 50:50.[7]
Permeability
[edit]The inner membrane is freely permeable to oxygen, carbon dioxide, and water only.[8] It is much less permeable to ions and small molecules than the outer membrane, creating compartments by separating the matrix from the cytosolic environment. This compartmentalization is a necessary feature for metabolism. The inner mitochondrial membrane is both an electrical insulator and chemical barrier. Sophisticated ion transporters exist to allow specific molecules to cross this barrier. There are several antiport systems embedded in the inner membrane, allowing exchange of anions between the cytosol and the mitochondrial matrix.[7]
IMM-associated proteins
[edit]- ATP–ADP translocase
- ATP-binding cassette transporter
- Cholesterol side-chain cleavage enzyme
- Protein tyrosine phosphatase
- Carnitine O-palmitoyltransferase
- Carnitine O-acetyltransferase
- Carnitine O-octanoyltransferase
- Cytochrome P450
- Translocase of the inner membrane
- Glutamate aspartate transporter
- Pyrimidine metabolism
See also
[edit]- Citric acid cycle
- Proton gradient
- Mitochondrial trifunctional protein
- Mitochondrial shuttle
- Transport proteins
References
[edit]- ^ Mannella CA (2006). "Structure and dynamics of the mitochondrial inner membrane cristae". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1763 (5–6): 542–548. doi:10.1016/j.bbamcr.2006.04.006. PMID 16730811.
- ^ Herrmann, JM (18 October 2011). "MINOS is plus: a Mitofilin complex for mitochondrial membrane contacts". Developmental Cell. 21 (4): 599–600. doi:10.1016/j.devcel.2011.09.013. PMID 22014515.
- ^ von der Malsburg, K; Müller, JM; Bohnert, M; Oeljeklaus, S; Kwiatkowska, P; Becker, T; Loniewska-Lwowska, A; Wiese, S; Rao, S; Milenkovic, D; Hutu, DP; Zerbes, RM; Schulze-Specking, A; Meyer, HE; Martinou, JC; Rospert, S; Rehling, P; Meisinger, C; Veenhuis, M; Warscheid, B; van der Klei, IJ; Pfanner, N; Chacinska, A; van der Laan, M (18 October 2011). "Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis" (PDF). Developmental Cell. 21 (4): 694–707. doi:10.1016/j.devcel.2011.08.026. PMID 21944719.
- ^ Rabl, R; Soubannier, V; Scholz, R; Vogel, F; Mendl, N; Vasiljev-Neumeyer, A; Körner, C; Jagasia, R; Keil, T; Baumeister, W; Cyrklaff, M; Neupert, W; Reichert, AS (15 June 2009). "Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g". The Journal of Cell Biology. 185 (6): 1047–63. doi:10.1083/jcb.200811099. PMC 2711607. PMID 19528297.
- ^ Comte J, Maïsterrena B, Gautheron DC (January 1976). "Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes". Biochim. Biophys. Acta. 419 (2): 271–84. doi:10.1016/0005-2736(76)90353-9. PMID 1247555.
- ^ Lomize, Andrel; Lomize, Mikhail; Pogozheva, Irina (2013). "Membrane Protein Lipid Composition Atlas". Orientations of Proteins in Membranes. University of Michigan. Retrieved 10 April 2014.
- ^ a b Krauss, Stefan (2001). "Mitochondria: Structure and Role in Respiration" (PDF). Nature Publishing Group. Archived from the original (PDF) on 21 October 2012. Retrieved 9 April 2014.
- ^ Caprette, David R. (12 December 1996). "Structure of Mitochondria". Experimental Biosciences. Rice University. Retrieved 9 April 2014.
External links
[edit]- Mitochondrial inner membrane (97 proteins) Orientations of Proteins in Membranes (OPM) database