Green fluorescent protein
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
| Green fluorescent protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
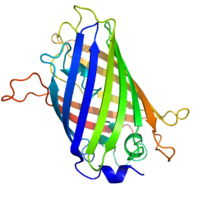
 Structure of the Aequorea victoria green fluorescent protein.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | GFP | ||||||||
| Pfam | PF01353 | ||||||||
| Pfam clan | CL0069 | ||||||||
| InterPro | IPR011584 | ||||||||
| CATH | 1ema | ||||||||
| SCOP2 | 1ema / SCOPe / SUPFAM | ||||||||
| |||||||||
| Green fluorescent protein | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | GFP | ||||||
| UniProt | P42212 | ||||||
| |||||||
The green fluorescent protein (GFP) is a protein that exhibits green fluorescence when exposed to light in the blue to ultraviolet range.[2][3] The label GFP traditionally refers to the protein first isolated from the jellyfish Aequorea victoria and is sometimes called avGFP. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets.[4]
The GFP from A. victoria has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (Renilla reniformis) has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other than molecular oxygen.[5]
In cell and molecular biology, the GFP gene is frequently used as a reporter of expression.[6] It has been used in modified forms to make biosensors, and many animals have been created that express GFP, which demonstrates a proof of concept that a gene can be expressed throughout a given organism, in selected organs, or in cells of interest. GFP can be introduced into animals or other species through transgenic techniques, and maintained in their genome and that of their offspring. GFP has been expressed in many species, including bacteria, yeasts, fungi, fish and mammals, including in human cells. Scientists Roger Y. Tsien, Osamu Shimomura, and Martin Chalfie were awarded the 2008 Nobel Prize in Chemistry on 10 October 2008 for their discovery and development of the green fluorescent protein.
Most commercially available genes for GFP and similar fluorescent proteins are around 730 base-pairs long. The natural protein has 238 amino acids. Its molecular mass is 27 kD.[7] Therefore, fusing the GFP gene to the gene of a protein of interest can significantly increase the protein's size and molecular mass, and can impair the protein's natural function or change its location or trajectory of transport within the cell.[8]
Background
[edit]

Wild-type GFP (wtGFP)
[edit]In the 1960s and 1970s, GFP, along with the separate luminescent protein aequorin (an enzyme that catalyzes the breakdown of luciferin, releasing light), was first purified from the jellyfish Aequorea victoria and its properties studied by Osamu Shimomura.[9] In A. victoria, GFP fluorescence occurs when aequorin interacts with Ca2+ ions, inducing a blue glow. Some of this luminescent energy is transferred to the GFP, shifting the overall color towards green.[10] However, its utility as a tool for molecular biologists did not begin to be realized until 1992 when Douglas Prasher reported the cloning and nucleotide sequence of wtGFP in Gene.[11] The funding for this project had run out, so Prasher sent cDNA samples to several labs. The lab of Martin Chalfie expressed the coding sequence of wtGFP, with the first few amino acids deleted, in heterologous cells of E. coli and C. elegans, publishing the results in Science in 1994.[12] Frederick Tsuji's lab independently reported the expression of the recombinant protein one month later.[13] Remarkably, the GFP molecule folded and was fluorescent at room temperature, without the need for exogenous cofactors specific to the jellyfish. Although this near-wtGFP was fluorescent, it had several drawbacks, including dual peaked excitation spectra, pH sensitivity, chloride sensitivity, poor fluorescence quantum yield, poor photostability and poor folding at 37 °C (99 °F).
The first reported crystal structure of a GFP was that of the S65T mutant by the Remington group in Science in 1996.[14] One month later, the Phillips group independently reported the wild-type GFP structure in Nature Biotechnology.[15] These crystal structures provided vital background on chromophore formation and neighboring residue interactions. Researchers have modified these residues by directed and random mutagenesis to produce the wide variety of GFP derivatives in use today. Further research into GFP has shown that it is resistant to detergents, proteases, guanidinium chloride (GdmCl) treatments, and drastic temperature changes.[16]
GFP derivatives
[edit]
Due to the potential for widespread usage and the evolving needs of researchers, many different mutants of GFP have been engineered.[17][18] The first major improvement was a single point mutation (S65T) reported in 1995 in Nature by Roger Tsien.[19] This mutation dramatically improved the spectral characteristics of GFP, resulting in increased fluorescence, photostability, and a shift of the major excitation peak to 488 nm, with the peak emission kept at 509 nm. This matched the spectral characteristics of commonly available FITC filter sets, increasing the practicality of use by the general researcher. A 37 °C folding efficiency (F64L) point mutant to this scaffold, yielding enhanced GFP (EGFP), was discovered in 1995 by the laboratories of Thastrup[20] and Falkow.[21] EGFP allowed the practical use of GFPs in mammalian cells. EGFP has an extinction coefficient (denoted ε) of 55,000 M−1cm−1.[22] The fluorescence quantum yield (QY) of EGFP is 0.60. The relative brightness, expressed as ε•QY, is 33,000 M−1cm−1.
Superfolder GFP (sfGFP), a series of mutations that allow GFP to rapidly fold and mature even when fused to poorly folding peptides, was reported in 2006.[23]
Many other mutations have been made, including color mutants; in particular, blue fluorescent protein (EBFP, EBFP2, Azurite, mKalama1), cyan fluorescent protein (ECFP, Cerulean, CyPet, mTurquoise2), and yellow fluorescent protein derivatives (YFP, Citrine, Venus, YPet). BFP derivatives (except mKalama1) contain the Y66H substitution. They exhibit a broad absorption band in the ultraviolet centered close to 380 nanometers and an emission maximum at 448 nanometers. A green fluorescent protein mutant (BFPms1) that preferentially binds Zn(II) and Cu(II) has been developed. BFPms1 have several important mutations including and the BFP chromophore (Y66H),Y145F for higher quantum yield, H148G for creating a hole into the beta-barrel and several other mutations that increase solubility. Zn(II) binding increases fluorescence intensity, while Cu(II) binding quenches fluorescence and shifts the absorbance maximum from 379 to 444 nm. Therefore, they can be used as Zn biosensor.[24]

Chromophore binding. The critical mutation in cyan derivatives is the Y66W substitution, which causes the chromophore to form with an indole rather than phenol component. Several additional compensatory mutations in the surrounding barrel are required to restore brightness to this modified chromophore due to the increased bulk of the indole group. In ECFP and Cerulean, the N-terminal half of the seventh strand exhibits two conformations. These conformations both have a complex set of van der Waals interactions with the chromophore. The Y145A and H148D mutations in Cerulean stabilize these interactions and allow the chromophore to be more planar, better packed, and less prone to collisional quenching.[25]
Additional site-directed random mutagenesis in combination with fluorescence lifetime based screening has further stabilized the seventh β-strand resulting in a bright variant, mTurquoise2, with a quantum yield (QY) of 0.93.[26] The red-shifted wavelength of the YFP derivatives is accomplished by the T203Y mutation and is due to π-electron stacking interactions between the substituted tyrosine residue and the chromophore.[3] These two classes of spectral variants are often employed for Förster resonance energy transfer (FRET) experiments. Genetically encoded FRET reporters sensitive to cell signaling molecules, such as calcium or glutamate, protein phosphorylation state, protein complementation, receptor dimerization, and other processes provide highly specific optical readouts of cell activity in real time.
Semirational mutagenesis of a number of residues led to pH-sensitive mutants known as pHluorins, and later super-ecliptic pHluorins. By exploiting the rapid change in pH upon synaptic vesicle fusion, pHluorins tagged to synaptobrevin have been used to visualize synaptic activity in neurons.[27]
Redox sensitive GFP (roGFP) was engineered by introduction of cysteines into the beta barrel structure. The redox state of the cysteines determines the fluorescent properties of roGFP.[28]
Nomenclature
[edit]The nomenclature of modified GFPs is often confusing due to overlapping mapping of several GFP versions onto a single name. For example, mGFP often refers to a GFP with an N-terminal palmitoylation that causes the GFP to bind to cell membranes. However, the same term is also used to refer to monomeric GFP, which is often achieved by the dimer interface breaking A206K mutation.[29] Wild-type GFP has a weak dimerization tendency at concentrations above 5 mg/mL. mGFP also stands for "modified GFP," which has been optimized through amino acid exchange for stable expression in plant cells.
In nature
[edit]

The purpose of both the (primary) bioluminescence (from aequorin's action on luciferin) and the (secondary) fluorescence of GFP in jellyfish is unknown. GFP is co-expressed with aequorin in small granules around the rim of the jellyfish bell. The secondary excitation peak (480 nm) of GFP does absorb some of the blue emission of aequorin, giving the bioluminescence a more green hue. The serine 65 residue of the GFP chromophore is responsible for the dual-peaked excitation spectra of wild-type GFP. It is conserved in all three GFP isoforms originally cloned by Prasher. Nearly all mutations of this residue consolidate the excitation spectra to a single peak at either 395 nm or 480 nm. The precise mechanism of this sensitivity is complex, but, it seems, involves donation of a hydrogen from serine 65 to glutamate 222, which influences chromophore ionization.[3] Since a single mutation can dramatically enhance the 480 nm excitation peak, making GFP a much more efficient partner of aequorin, A. victoria appears to evolutionarily prefer the less-efficient, dual-peaked excitation spectrum. Roger Tsien has speculated that varying hydrostatic pressure with depth may affect serine 65's ability to donate a hydrogen to the chromophore and shift the ratio of the two excitation peaks. Thus, the jellyfish may change the color of its bioluminescence with depth. However, a collapse in the population of jellyfish in Friday Harbor, where GFP was originally discovered, has hampered further study of the role of GFP in the jellyfish's natural environment.
Most species of lancelet are known to produce GFP in various regions of their body.[30] Unlike A. victoria, lancelets do not produce their own blue light, and the origin of their endogenous GFP is still unknown. Some speculate that it attracts plankton towards the mouth of the lancelet, serving as a passive hunting mechanism. It may also serve as a photoprotective agent in the larvae, preventing damage caused by high-intensity blue light by converting it into lower-intensity green light. However, these theories have not been tested.
GFP-like proteins have been found in multiple species of marine copepods, particularly from the Pontellidae and Aetideidae families.[31] GFP isolated from Pontella mimocerami has shown high levels of brightness with a quantum yield of 0.92, making them nearly two-fold brighter than the commonly used EGFP isolated from A. victoria.[32]
Other fluorescent proteins
[edit]
There are many GFP-like proteins that, despite being in the same protein family as GFP, are not directly derived from Aequorea victoria. These include dsRed, eqFP611, Dronpa, TagRFPs, KFP, EosFP/IrisFP, Dendra, and so on. Having been developed from proteins in different organisms, these proteins can sometimes display unanticipated approaches to chromophore formation. Some of these, such as KFP, are developed from naturally non- or weakly-fluorescent proteins to be greatly improved upon by mutagenesis.[33] When GFP-like barrels of different spectra characteristics are used, the excitation spectra of one chromophore can be used to power another chromophore (FRET), allowing for conversion between wavelengths of light.[34]
FMN-binding fluorescent proteins (FbFPs) were developed in 2007 and are a class of small (11–16 kDa), oxygen-independent fluorescent proteins that are derived from blue-light receptors. They are intended especially for the use under anaerobic or hypoxic conditions, since the formation and binding of the Flavin chromophore does not require molecular oxygen, as it is the case with the synthesis of the GFP chromophore.[35]

Fluorescent proteins with other chromophores, such as UnaG with bilirubin, can display unique properties like red-shifted emission above 600 nm or photoconversion from a green-emitting state to a red-emitting state. They can have excitation and emission wavelengths far enough apart to achieve conversion between red and green light.
A new class of fluorescent protein was evolved from a cyanobacterial (Trichodesmium erythraeum) phycobiliprotein, α-allophycocyanin, and named small ultra red fluorescent protein (smURFP) in 2016. smURFP autocatalytically self-incorporates the chromophore biliverdin without the need of an external protein, known as a lyase.[36][37] Jellyfish- and coral-derived GFP-like proteins require oxygen and produce a stoichiometric amount of hydrogen peroxide upon chromophore formation.[38] smURFP does not require oxygen or produce hydrogen peroxide and uses the chromophore, biliverdin. smURFP has a large extinction coefficient (180,000 M−1 cm−1) and has a modest quantum yield (0.20), which makes it comparable biophysical brightness to eGFP and ~2-fold brighter than most red or far-red fluorescent proteins derived from coral. smURFP spectral properties are similar to the organic dye Cy5.[36][39]

Reviews on new classes of fluorescent proteins and applications can be found in the cited reviews.[40][41]
Structure
[edit]GFP has a beta barrel structure consisting of eleven β-strands with a pleated sheet arrangement, with an alpha helix containing the covalently bonded chromophore 4-(p-hydroxybenzylidene)imidazolidin-5-one (HBI) running through the center.[3][14][15] Five shorter alpha helices form caps on the ends of the structure. The beta barrel structure is a nearly perfect cylinder, 42Å long and 24Å in diameter (some studies have reported a diameter of 30Å[16]),[14] creating what is referred to as a "β-can" formation, which is unique to the GFP-like family.[15] HBI, the spontaneously modified form of the tripeptide Ser65–Tyr66–Gly67, is nonfluorescent in the absence of the properly folded GFP scaffold and exists mainly in the un-ionized phenol form in wtGFP.[42] Inward-facing sidechains of the barrel induce specific cyclization reactions in Ser65–Tyr66–Gly67 that induce ionization of HBI to the phenolate form and chromophore formation. This process of post-translational modification is referred to as maturation.[43] The hydrogen-bonding network and electron-stacking interactions with these sidechains influence the color, intensity and photostability of GFP and its numerous derivatives.[44] The tightly packed nature of the barrel excludes solvent molecules, protecting the chromophore fluorescence from quenching by water. In addition to the auto-cyclization of the Ser65-Tyr66-Gly67, a 1,2-dehydrogenation reaction occurs at the Tyr66 residue.[16] Besides the three residues that form the chromophore, residues such as Gln94, Arg96, His148, Thr203, and Glu222 all act as stabilizers. The residues of Gln94, Arg96, and His148 are able to stabilize by delocalizing the chromophore charge. Arg96 is the most important stabilizing residue due to the fact that it prompts the necessary structural realignments that are necessary from the HBI ring to occur. Any mutation to the Arg96 residue would result in a decrease in the development rate of the chromophore because proper electrostatic and steric interactions would be lost. Tyr66 is the recipient of hydrogen bonds and does not ionize in order to produce favorable electrostatics.[45]

 |  |
Autocatalytic formation of the chromophore in wtGFP
[edit]Mechanistically, the process involves base-mediated cyclization followed by dehydration and oxidation. In the reaction of 7a to 8 involves the formation of an enamine from the imine, while in the reaction of 7b to 9 a proton is abstracted.[46] The formed HBI fluorophore is highlighted in green.
The reactions are catalyzed by residues Glu222 and Arg96.[46][47] An analogous mechanism is also possible with threonine in place of Ser65.
Applications
[edit]Reporter assays
[edit]Green fluorescent protein may be used as a reporter gene.[48][49]
For example, GFP can be used as a reporter for environmental toxicity levels. This protein has been shown to be an effective way to measure the toxicity levels of various chemicals including ethanol, p-formaldehyde, phenol, triclosan, and paraben. GFP is great as a reporter protein because it has no effect on the host when introduced to the host's cellular environment. Due to this ability, no external visualization stain, ATP, or cofactors are needed. With regards to pollutant levels, the fluorescence was measured in order to gauge the effect that the pollutants have on the host cell. The cellular density of the host cell was also measured. Results from the study conducted by Song, Kim, & Seo (2016) showed that there was a decrease in both fluorescence and cellular density as pollutant levels increased. This was indicative of the fact that cellular activity had decreased. More research into this specific application in order to determine the mechanism by which GFP acts as a pollutant marker.[50] Similar results have been observed in zebrafish because zebrafish that were injected with GFP were approximately twenty times more susceptible to recognize cellular stresses than zebrafish that were not injected with GFP.[51]
Advantages
[edit]The biggest advantage of GFP is that it can be heritable, depending on how it was introduced, allowing for continued study of cells and tissues it is expressed in. Visualizing GFP is noninvasive, requiring only illumination with blue light. GFP alone does not interfere with biological processes, but when fused to proteins of interest, careful design of linkers is required to maintain the function of the protein of interest. Moreover, if used with a monomer it is able to diffuse readily throughout cells.[52]
Fluorescence microscopy
[edit]
The availability of GFP and its derivatives has thoroughly redefined fluorescence microscopy and the way it is used in cell biology and other biological disciplines.[53] While most small fluorescent molecules such as FITC (fluorescein isothiocyanate) are strongly phototoxic when used in live cells, fluorescent proteins such as GFP are usually much less harmful when illuminated in living cells. This has triggered the development of highly automated live-cell fluorescence microscopy systems, which can be used to observe cells over time expressing one or more proteins tagged with fluorescent proteins.
There are many techniques to utilize GFP in a live cell imaging experiment. The most direct way of utilizing GFP is to directly attach it to a protein of interest. For example, GFP can be included in a plasmid expressing other genes to indicate a successful transfection of a gene of interest. Another method is to use a GFP that contains a mutation where the fluorescence will change from green to yellow over time, which is referred to as a fluorescent timer. With the fluorescent timer, researchers can study the state of protein production such as recently activated, continuously activated, or recently deactivated based on the color reported by the fluorescent protein.[54] In yet another example, scientists have modified GFP to become active only after exposure to irradiation giving researchers a tool to selectively activate certain portions of a cell and observe where proteins tagged with the GFP move from the starting location.[55] These are only two examples in a burgeoning field of fluorescent microcopy and a more complete review of biosensors utilizing GFP and other fluorescent proteins can be found here [56]
For example, GFP had been widely used in labelling the spermatozoa of various organisms for identification purposes as in Drosophila melanogaster, where expression of GFP can be used as a marker for a particular characteristic. GFP can also be expressed in different structures enabling morphological distinction. In such cases, the gene for the production of GFP is incorporated into the genome of the organism in the region of the DNA that codes for the target proteins and that is controlled by the same regulatory sequence; that is, the gene's regulatory sequence now controls the production of GFP, in addition to the tagged protein(s). In cells where the gene is expressed, and the tagged proteins are produced, GFP is produced at the same time. Thus, only those cells in which the tagged gene is expressed, or the target proteins are produced, will fluoresce when observed under fluorescence microscopy. Analysis of such time lapse movies has redefined the understanding of many biological processes including protein folding, protein transport, and RNA dynamics, which in the past had been studied using fixed (i.e., dead) material. Obtained data are also used to calibrate mathematical models of intracellular systems and to estimate rates of gene expression.[57] Similarly, GFP can be used as an indicator of protein expression in heterologous systems. In this scenario, fusion proteins containing GFP are introduced indirectly, using RNA of the construct, or directly, with the tagged protein itself. This method is useful for studying structural and functional characteristics of the tagged protein on a macromolecular or single-molecule scale with fluorescence microscopy.
The Vertico SMI microscope using the SPDM Phymod technology uses the so-called "reversible photobleaching" effect of fluorescent dyes like GFP and its derivatives to localize them as single molecules in an optical resolution of 10 nm. This can also be performed as a co-localization of two GFP derivatives (2CLM).[58]
Another powerful use of GFP is to express the protein in small sets of specific cells. This allows researchers to optically detect specific types of cells in vitro (in a dish), or even in vivo (in the living organism).[59] Genetically combining several spectral variants of GFP is a useful trick for the analysis of brain circuitry (Brainbow).[60] Other interesting uses of fluorescent proteins in the literature include using FPs as sensors of neuron membrane potential,[61] tracking of AMPA receptors on cell membranes,[62] viral entry and the infection of individual influenza viruses and lentiviral viruses,[63][64] etc.
It has also been found that new lines of transgenic GFP rats can be relevant for gene therapy as well as regenerative medicine.[65] By using "high-expresser" GFP, transgenic rats display high expression in most tissues, and many cells that have not been characterized or have been only poorly characterized in previous GFP-transgenic rats.
GFP has been shown to be useful in cryobiology as a viability assay. Correlation of viability as measured by trypan blue assays were 0.97.[66] Another application is the use of GFP co-transfection as internal control for transfection efficiency in mammalian cells.[67]
A novel possible use of GFP includes using it as a sensitive monitor of intracellular processes via an eGFP laser system made out of a human embryonic kidney cell line. The first engineered living laser is made by an eGFP expressing cell inside a reflective optical cavity and hitting it with pulses of blue light. At a certain pulse threshold, the eGFP's optical output becomes brighter and completely uniform in color of pure green with a wavelength of 516 nm. Before being emitted as laser light, the light bounces back and forth within the resonator cavity and passes the cell numerous times. By studying the changes in optical activity, researchers may better understand cellular processes.[68][69]
GFP is used widely in cancer research to label and track cancer cells. GFP-labelled cancer cells have been used to model metastasis, the process by which cancer cells spread to distant organs.[70]
Split GFP
[edit]GFP can be used to analyse the colocalization of proteins. This is achieved by "splitting" the protein into two fragments which are able to self-assemble, and then fusing each of these to the two proteins of interest. Alone, these incomplete GFP fragments are unable to fluoresce. However, if the two proteins of interest colocalize, then the two GFP fragments assemble together to form a GFP-like structure which is able to fluoresce. Therefore, by measuring the level of fluorescence it is possible to determine whether the two proteins of interest colocalize.[71]
Macro-photography
[edit]Macro-scale biological processes, such as the spread of virus infections, can be followed using GFP labeling.[72] In the past, mutagenic ultra violet light (UV) has been used to illuminate living organisms (e.g., see[73]) to detect and photograph the GFP expression. Recently, a technique using non-mutagenic LED lights[74] have been developed for macro-photography.[75] The technique uses an epifluorescence camera attachment[76] based on the same principle used in the construction of epifluorescence microscopes.
Transgenic pets
[edit]
Alba, a green-fluorescent rabbit, was created by a French laboratory commissioned by Eduardo Kac using GFP for purposes of art and social commentary.[77] The US company Yorktown Technologies markets to aquarium shops green fluorescent zebrafish (GloFish) that were initially developed to detect pollution in waterways. NeonPets, a US-based company has marketed green fluorescent mice to the pet industry as NeonMice.[78] Green fluorescent pigs, known as Noels, were bred by a group of researchers led by Wu Shinn-Chih at the Department of Animal Science and Technology at National Taiwan University.[79] A Japanese-American Team created green-fluorescent cats as proof of concept to use them potentially as model organisms for diseases, particularly HIV.[80] In 2009 a South Korean team from Seoul National University bred the first transgenic beagles with fibroblast cells from sea anemones. The dogs give off a red fluorescent light, and they are meant to allow scientists to study the genes that cause human diseases like narcolepsy and blindness.[81]
Art
[edit]Julian Voss-Andreae, a German-born artist specializing in "protein sculptures,"[82] created sculptures based on the structure of GFP, including the 1.70 metres (5 feet 7 inches) tall "Green Fluorescent Protein" (2004)[83] and the 1.40 metres (4 feet 7 inches) tall "Steel Jellyfish" (2006). The latter sculpture is located at the place of GFP's discovery by Shimomura in 1962, the University of Washington's Friday Harbor Laboratories.[84]

See also
[edit]References
[edit]- ^ Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (September 1996). "Crystal structure of the Aequorea victoria green fluorescent protein". Science. 273 (5280): 1392–5. Bibcode:1996Sci...273.1392O. doi:10.1126/science.273.5280.1392. PMID 8703075. S2CID 43030290.
- ^ Prendergast FG, Mann KG (Aug 1978). "Chemical and physical properties of aequorin and the green fluorescent protein isolated from Aequorea forskålea". Biochemistry. 17 (17): 3448–53. doi:10.1021/bi00610a004. PMID 28749.
- ^ a b c d Tsien RY (1998). "The green fluorescent protein" (PDF). Annual Review of Biochemistry. 67: 509–44. doi:10.1146/annurev.biochem.67.1.509. PMID 9759496.
- ^ Salih A (2019). "Fluorescent Proteins". In Cox G (ed.). Fundamentals of Fluorescence Imaging. Boca Raton: Jenny Stanford Publishing. p. 122. doi:10.1201/9781351129404. ISBN 9781351129404. S2CID 213688192.
- ^ Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK (Aug 2008). "Fluorescent proteins as biomarkers and biosensors: throwing color lights on molecular and cellular processes". Current Protein & Peptide Science. 9 (4): 338–69. doi:10.2174/138920308785132668. PMC 2904242. PMID 18691124.
- ^ Phillips GJ (Oct 2001). "Green fluorescent protein—a bright idea for the study of bacterial protein localization". FEMS Microbiology Letters. 204 (1): 9–18. doi:10.1016/S0378-1097(01)00358-5. PMID 11682170.
- ^ Uckert W, Pedersen L, Günzburg W (2000). "Green fluorescent protein retroviral vector: generation of high-titer producer cells and virus supernatant". Gene Therapy of Cancer. Methods in Molecular Medicine. Vol. 35. pp. 275–85. doi:10.1385/1-59259-086-1:275. ISBN 1-59259-086-1. PMID 21390811.
- ^ Goodman SR, ed. (2008). "Chapter 1 - Tools of the Cell Biologist: Green Fluorescent Protein". Medical Cell Biology (3rd ed.). Amsterdam: Elsevier/Academic Press. pp. 14–25. doi:10.1016/B978-0-12-370458-0.50006-2. ISBN 978-0-12-370458-0. S2CID 90224559.
- ^ Shimomura O, Johnson FH, Saiga Y (Jun 1962). "Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea". Journal of Cellular and Comparative Physiology. 59 (3): 223–39. doi:10.1002/jcp.1030590302. PMID 13911999.
- ^ Morise H, Shimomura O, Johnson FH, Winant J (Jun 1974). "Intermolecular energy transfer in the bioluminescent system of Aequorea". Biochemistry. 13 (12): 2656–62. doi:10.1021/bi00709a028. PMID 4151620.
- ^ Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ (Feb 1992). "Primary structure of the Aequorea victoria green-fluorescent protein". Gene. 111 (2): 229–33. doi:10.1016/0378-1119(92)90691-H. PMID 1347277.
- ^ Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (Feb 1994). "Green fluorescent protein as a marker for gene expression". Science. 263 (5148): 802–5. Bibcode:1994Sci...263..802C. doi:10.1126/science.8303295. PMID 8303295. S2CID 9043327.
- ^ Inouye S, Tsuji FI (Mar 1994). "Aequorea green fluorescent protein. Expression of the gene and fluorescence characteristics of the recombinant protein". FEBS Letters. 341 (2–3): 277–80. doi:10.1016/0014-5793(94)80472-9. PMID 8137953.
- ^ a b c Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (Sep 1996). "Crystal structure of the Aequorea victoria green fluorescent protein". Science. 273 (5280): 1392–5. Bibcode:1996Sci...273.1392O. doi:10.1126/science.273.5280.1392. PMID 8703075. S2CID 43030290.
- ^ a b c Yang F, Moss LG, Phillips GN (Oct 1996). "The molecular structure of green fluorescent protein" (PDF). Nature Biotechnology. 14 (10): 1246–51. doi:10.1038/nbt1096-1246. hdl:1911/19233. PMID 9631087. S2CID 34713931.
- ^ a b c Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormö M, Remington SJ (March 1997). "Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein". Proceedings of the National Academy of Sciences of the United States of America. 94 (6): 2306–2311. Bibcode:1997PNAS...94.2306B. doi:10.1073/pnas.94.6.2306. PMC 20083. PMID 9122190.
- ^ Shaner NC, Steinbach PA, Tsien RY (Dec 2005). "A guide to choosing fluorescent proteins" (PDF). Nature Methods. 2 (12): 905–9. doi:10.1038/nmeth819. PMID 16299475. S2CID 10024284.
- ^ Wilhelmsson M, Tor Y (2016). Fluorescent Analogs of Biomolecular Building Blocks: Design and Applications. New Jersey: Wiley. ISBN 978-1-118-17586-6.
- ^ Heim R, Cubitt AB, Tsien RY (Feb 1995). "Improved green fluorescence" (PDF). Nature. 373 (6516): 663–4. Bibcode:1995Natur.373..663H. doi:10.1038/373663b0. PMID 7854443. S2CID 40179694.
- ^ US patent 6172188, Thastrup O, Tullin S, Kongsbak Poulsen L, Bjørn S, "Fluorescent Proteins", published 2001-01-09
- ^ Cormack BP, Valdivia RH, Falkow S (1996). "FACS-optimized mutants of the green fluorescent protein (GFP)". Gene. 173 (1 Spec No): 33–38. doi:10.1016/0378-1119(95)00685-0. PMID 8707053.
- ^ McRae SR, Brown CL, Bushell GR (May 2005). "Rapid purification of EGFP, EYFP, and ECFP with high yield and purity". Protein Expression and Purification. 41 (1): 121–127. doi:10.1016/j.pep.2004.12.030. PMID 15802229.
- ^ Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (Jan 2006). "Engineering and characterization of a superfolder green fluorescent protein". Nature Biotechnology. 24 (1): 79–88. doi:10.1038/nbt1172. PMID 16369541. S2CID 2966399.
- ^ Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED (Apr 2002). "Structural chemistry of a green fluorescent protein Zn biosensor". Journal of the American Chemical Society. 124 (14): 3522–3524. doi:10.1021/ja0176954. PMID 11929238.
- ^ Lelimousin M, Noirclerc-Savoye M, Lazareno-Saez C, Paetzold B, Le Vot S, Chazal R, Macheboeuf P, Field MJ, Bourgeois D, Royant A (Oct 2009). "Intrinsic dynamics in ECFP and Cerulean control fluorescence quantum yield". Biochemistry. 48 (42): 10038–10046. doi:10.1021/bi901093w. PMID 19754158.
- ^ Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW, Royant A (2012). "Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%". Nature Communications. 3: 751. Bibcode:2012NatCo...3..751G. doi:10.1038/ncomms1738. PMC 3316892. PMID 22434194.
- ^ Miesenböck G, De Angelis DA, Rothman JE (Jul 1998). "Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins". Nature. 394 (6689): 192–5. Bibcode:1998Natur.394..192M. doi:10.1038/28190. PMID 9671304. S2CID 4320849.
- ^ Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ (Mar 2004). "Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators". The Journal of Biological Chemistry. 279 (13): 13044–53. doi:10.1074/jbc.M312846200. PMID 14722062.
- ^ Zacharias DA, Violin JD, Newton AC, Tsien RY (May 2002). "Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells". Science. 296 (5569): 913–16. Bibcode:2002Sci...296..913Z. doi:10.1126/science.1068539. PMID 11988576. S2CID 14957077.
- ^ Yue JX, Holland ND, Holland LZ, Deheyn DD (June 2016). "The evolution of genes encoding for green fluorescent proteins: insights from cephalochordates (amphioxus)". Scientific Reports. 6 (1): 28350. Bibcode:2016NatSR...628350Y. doi:10.1038/srep28350. PMC 4911609. PMID 27311567.
- ^ Hunt ME, Scherrer MP, Ferrari FD, Matz MV (July 2010). "Very bright green fluorescent proteins from the Pontellid copepod Pontella mimocerami". PLOS ONE. 5 (7): e11517. Bibcode:2010PLoSO...511517H. doi:10.1371/journal.pone.0011517. PMC 2904364. PMID 20644720.
- ^ "eGFP". FPbase.
- ^ Chudakov DM, Belousov VV, Zaraisky AG, Novoselov VV, Staroverov DB, Zorov DB, Lukyanov S, Lukyanov KA (February 2003). "Kindling fluorescent proteins for precise in vivo photolabeling". Nature Biotechnology. 21 (2): 191–4. doi:10.1038/nbt778. PMID 12524551. S2CID 52887792.
- ^ Wiens MD, Shen Y, Li X, Salem MA, Smisdom N, Zhang W, Brown A, Campbell RE (December 2016). "A Tandem Green-Red Heterodimeric Fluorescent Protein with High FRET Efficiency". ChemBioChem. 17 (24): 2361–2367. doi:10.1002/cbic.201600492. PMID 27781394. S2CID 4301322.
- ^ Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl JK, et al. (April 2007). "Reporter proteins for in vivo fluorescence without oxygen". Nature Biotechnology. 25 (4): 443–445. doi:10.1038/nbt1293. PMID 17351616. S2CID 7335755.
- ^ a b Rodriguez EA, Tran GN, Gross LA, Crisp JL, Shu X, Lin JY, Tsien RY (September 2016). "A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein". Nature Methods. 13 (9): 763–9. doi:10.1038/nmeth.3935. PMC 5007177. PMID 27479328.
- ^ Mattson S, Tran GN, Rodriguez EA (2023). "Directed Evolution of Fluorescent Proteins in Bacteria". In Sharma M (ed.). Fluorescent Proteins. Methods in Molecular Biology. Vol. 2564. New York, NY: Springer US. pp. 75–97. doi:10.1007/978-1-0716-2667-2_4. ISBN 978-1-0716-2666-5. PMID 36107338.
- ^ Tsien RY (1998-01-01). "The green fluorescent protein". Annual Review of Biochemistry. 67 (1): 509–44. doi:10.1146/annurev.biochem.67.1.509. PMID 9759496. S2CID 8138960.
- ^ Maiti, Atanu; Buffalo, Cosmo Z.; Saurabh, Saumya; Montecinos-Franjola, Felipe; Hachey, Justin S.; Conlon, William J.; Tran, Geraldine N.; Hassan, Bakar; Walters, Kylie J.; Drobizhev, Mikhail; Moerner, W. E.; Ghosh, Partho; Matsuo, Hiroshi; Tsien, Roger Y.; Lin, John Y. (2023-07-12). "Structural and photophysical characterization of the small ultra-red fluorescent protein". Nature Communications. 14 (1): 4155. Bibcode:2023NatCo..14.4155M. doi:10.1038/s41467-023-39776-9. ISSN 2041-1723. PMC 10338489. PMID 37438348.
- ^ Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, Tsien RY (February 2017). "The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins". Trends in Biochemical Sciences. 42 (2): 111–129. doi:10.1016/j.tibs.2016.09.010. PMC 5272834. PMID 27814948.
- ^ Montecinos-Franjola F, Lin JY, Rodriguez EA (2020-11-16). "Fluorescent proteins for in vivo imaging, where's the biliverdin?". Biochemical Society Transactions. 48 (6): 2657–2667. doi:10.1042/BST20200444. PMID 33196077. S2CID 226971864.
- ^ Bokman SH, Ward WW (1982). "Reversible denaturation of Aequorea green-fluorescent protein: physical separation and characterization of the renatured protein". Biochemistry. 21 (19): 4535–4540. doi:10.1021/bi00262a003. PMID 6128025.
- ^ Pouwels LJ, Zhang L, Chan NH, Dorrestein PC, Wachter RM (Sep 2008). "Kinetic isotope effect studies on the de novo rate of chromophore formation in fast- and slow-maturing GFP variants". Biochemistry. 47 (38): 10111–22. doi:10.1021/bi8007164. PMC 2643082. PMID 18759496.
- ^ Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA (Jul 2010). "Fluorescent proteins and their applications in imaging living cells and tissues". Physiological Reviews. 90 (3): 1103–63. doi:10.1152/physrev.00038.2009. PMID 20664080. S2CID 10767597.
- ^ Stepanenko OV, Verkhusha VV, Shavlovsky MM, Kuznetsova IM, Uversky VN, Turoverov KK (November 2008). "Understanding the role of Arg96 in structure and stability of green fluorescent protein". Proteins. 73 (3): 539–551. doi:10.1002/prot.22089. PMC 2908022. PMID 18470931.
- ^ a b Rosenow MA, Huffman HA, Phail ME, Wachter RM (April 2004). "The crystal structure of the Y66L variant of green fluorescent protein supports a cyclization-oxidation-dehydration mechanism for chromophore maturation". Biochemistry. 43 (15): 4464–4472. doi:10.1021/bi0361315. PMID 15078092.
- ^ Ma Y, Yu JG, Sun Q, Li Z, Smith SC (2015-07-01). "The mechanism of dehydration in chromophore maturation of wild-type green fluorescent protein: A theoretical study". Chemical Physics Letters. 631–632: 42–46. Bibcode:2015CPL...631...42M. doi:10.1016/j.cplett.2015.04.061. ISSN 0009-2614.
- ^ Jugder BE, Welch J, Braidy N, Marquis CP (2016-07-26). "Construction and use of a Cupriavidus necator H16 soluble hydrogenase promoter (PSH) fusion to gfp (green fluorescent protein)". PeerJ. 4: e2269. doi:10.7717/peerj.2269. PMC 4974937. PMID 27547572.
- ^ Arun KH, Kaul CL, Ramarao P (2005). "Green fluorescent proteins in receptor research: an emerging tool for drug discovery". Journal of Pharmacological and Toxicological Methods. 51 (1): 1–23. doi:10.1016/j.vascn.2004.07.006. PMID 15596111.
- ^ Song YH, Kim CS, Seo JH (April 2016). "Noninvasive monitoring of environmental toxicity through green fluorescent protein expressing Escherichia coli". Korean Journal of Chemical Engineering. 33 (4): 1331–6. doi:10.1007/s11814-015-0253-1. S2CID 62828580.
- ^ Pan Y, Leifert A, Graf M, Schiefer F, Thoröe-Boveleth S, Broda J, Halloran MC, Hollert H, Laaf D, Simon U, Jahnen-Dechent W (March 2013). "High-sensitivity real-time analysis of nanoparticle toxicity in green fluorescent protein-expressing zebrafish". Small. 9 (6). Weinheim an Der Bergstrasse, Germany: 863–9. doi:10.1002/smll.201201173. PMID 23143852.
- ^ Chalfie M (Jun 2009). "GFP: Lighting up life". Proceedings of the National Academy of Sciences of the United States of America. 106 (25): 10073–10080. Bibcode:2009PNAS..10610073C. doi:10.1073/pnas.0904061106. PMC 2700921. PMID 19553219.
- ^ Yuste R (Dec 2005). "Fluorescence microscopy today". Nature Methods. 2 (12): 902–4. doi:10.1038/nmeth1205-902. PMID 16299474. S2CID 205418407.
- ^ Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, et al. (November 2000). ""Fluorescent timer": protein that changes color with time". Science. 290 (5496): 1585–8. Bibcode:2000Sci...290.1585T. doi:10.1126/science.290.5496.1585. PMID 11090358.
- ^ Patterson GH, Lippincott-Schwartz J (September 2002). "A photoactivatable GFP for selective photolabeling of proteins and cells". Science. 297 (5588): 1873–7. Bibcode:2002Sci...297.1873P. doi:10.1126/science.1074952. PMID 12228718. S2CID 45058411.
- ^ Lin W, Mehta S, Zhang J (October 2019). "Genetically encoded fluorescent biosensors illuminate kinase signaling in cancer". The Journal of Biological Chemistry. 294 (40): 14814–14822. doi:10.1074/jbc.REV119.006177. PMC 6779441. PMID 31434714.
- ^ Komorowski M, Finkenstädt B, Rand D (Jun 2010). "Using a single fluorescent reporter gene to infer half-life of extrinsic noise and other parameters of gene expression". Biophysical Journal. 98 (12): 2759–2769. Bibcode:2010BpJ....98.2759K. doi:10.1016/j.bpj.2010.03.032. PMC 2884236. PMID 20550887.
- ^ Gunkel M, Erdel F, Rippe K, Lemmer P, Kaufmann R, Hörmann C, Amberger R, Cremer C (Jun 2009). "Dual color localization microscopy of cellular nanostructures". Biotechnology Journal. 4 (6): 927–38. doi:10.1002/biot.200900005. PMID 19548231. S2CID 18162278.
- ^ Chudakov DM, Lukyanov S, Lukyanov KA (Dec 2005). "Fluorescent proteins as a toolkit for in vivo imaging". Trends in Biotechnology. 23 (12): 605–13. doi:10.1016/j.tibtech.2005.10.005. PMID 16269193.
- ^ Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW (Nov 2007). "Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system". Nature. 450 (7166): 56–62. Bibcode:2007Natur.450...56L. doi:10.1038/nature06293. PMID 17972876. S2CID 4402093.
- ^ Baker BJ, Mutoh H, Dimitrov D, Akemann W, Perron A, Iwamoto Y, Jin L, Cohen LB, Isacoff EY, Pieribone VA, Hughes T, Knöpfel T (Aug 2008). "Genetically encoded fluorescent sensors of membrane potential". Brain Cell Biology. 36 (1–4): 53–67. doi:10.1007/s11068-008-9026-7. PMC 2775812. PMID 18679801.
- ^ Adesnik H, Nicoll RA, England PM (Dec 2005). "Photoinactivation of native AMPA receptors reveals their real-time trafficking". Neuron. 48 (6): 977–85. doi:10.1016/j.neuron.2005.11.030. PMID 16364901.
- ^ Lakadamyali M, Rust MJ, Babcock HP, Zhuang X (Aug 2003). "Visualizing infection of individual influenza viruses". Proceedings of the National Academy of Sciences of the United States of America. 100 (16): 9280–5. Bibcode:2003PNAS..100.9280L. doi:10.1073/pnas.0832269100. PMC 170909. PMID 12883000.
- ^ Joo KI, Wang P (Oct 2008). "Visualization of targeted transduction by engineered lentiviral vectors". Gene Therapy. 15 (20): 1384–96. doi:10.1038/gt.2008.87. PMC 2575058. PMID 18480844.
- ^ Remy S, Tesson L, Usal C, Menoret S, Bonnamain V, Nerriere-Daguin V, Rossignol J, Boyer C, Nguyen TH, Naveilhan P, Lescaudron L, Anegon I (Oct 2010). "New lines of GFP transgenic rats relevant for regenerative medicine and gene therapy". Transgenic Research. 19 (5): 745–63. doi:10.1007/s11248-009-9352-2. PMID 20094912. S2CID 42499768.
- ^ Elliott G, McGrath J, Crockett-Torabi E (Jun 2000). "Green fluorescent protein: A novel viability assay for cryobiological applications". Cryobiology. 40 (4): 360–369. doi:10.1006/cryo.2000.2258. PMID 10924267.
- ^ Fakhrudin N, Ladurner A, Atanasov AG, Heiss EH, Baumgartner L, Markt P, Schuster D, Ellmerer EP, Wolber G, Rollinger JM, Stuppner H, Dirsch VM (Apr 2010). "Computer-aided discovery, validation, and mechanistic characterization of novel neolignan activators of peroxisome proliferator-activated receptor gamma". Molecular Pharmacology. 77 (4): 559–66. doi:10.1124/mol.109.062141. PMC 3523390. PMID 20064974.
- ^ Gather MC, Yun SH (2011). "Single-cell biological lasers". Nature Photonics. 5 (7): 406–410. Bibcode:2011NaPho...5..406G. doi:10.1038/nphoton.2011.99. S2CID 54971962.
- ^ Matson J (2011). "Green Fluorescent Protein Makes for Living Lasers". Scientific American. Retrieved 2011-06-13.
- ^ Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, Pai SY, Ho IC, Werb Z (Feb 2008). "GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model". Cancer Cell. 13 (2): 141–52. doi:10.1016/j.ccr.2008.01.011. PMC 2262951. PMID 18242514.
- ^ Cabantous S, Terwilliger TC, Waldo GS (January 2005). "Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein". Nature Biotechnology. 23 (1): 102–7. doi:10.1038/nbt1044. PMID 15580262. S2CID 25833063.
- ^ Rodman MK, Yadav NS, Artus NN (2002-09-01). "Progression of geminivirus-induced transgene silencing is associated with transgene methylation". New Phytologist. 155 (3): 461–468. doi:10.1046/j.1469-8137.2002.00467.x. PMID 33873315.
- ^ Zhu YJ, Agbayani R, Moore PH (Apr 2004). "Green fluorescent protein as a visual selection marker for papaya (Carica papaya L.) transformation". Plant Cell Reports. 22 (9): 660–7. doi:10.1007/s00299-004-0755-5. PMID 14749892. S2CID 23198182.
- ^ Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999). "Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants". The Plant Journal. 18 (4): 455–63. doi:10.1046/j.1365-313X.1999.00464.x. PMID 10406127. S2CID 292648.
- ^ Baker SS, Vidican CB, Cameron DS, Greib HG, Jarocki CC, Setaputri AW, Spicuzza CH, Burr AA, Waqas MA, Tolbert DA (2012-01-01). "An epifluorescent attachment improves whole-plant digital photography of Arabidopsis thaliana expressing red-shifted green fluorescent protein". AoB Plants. 2012: pls003. doi:10.1093/aobpla/pls003. PMC 3296078. PMID 22479674.
- ^ "PlantEdDL - Using SRL digital cameras in quantitative investigations of plants expressing green fluorescent protein (GFP)". planted.botany.org. Retrieved 2016-03-23.
- ^ Kac E. "GFP Bunny".
- ^ "Glow-In-The Dark NeonMice". Archived from the original on February 14, 2009. Retrieved August 30, 2016.
- ^ Scientists in Taiwan breed fluorescent green pigs
- ^ Wongsrikeao P, Saenz D, Rinkoski T, Otoi T, Poeschla E (2011). "Antiviral restriction factor transgenesis in the domestic cat". Nature Methods. 8 (10): 853–9. doi:10.1038/nmeth.1703. PMC 4006694. PMID 21909101.
- ^ "Fluorescent puppy is world's first transgenic dog".
- ^ Voss-Andreae J (2005). "Protein Sculptures: Life's Building Blocks Inspire Art". Leonardo. 38: 41–45. doi:10.1162/leon.2005.38.1.41. S2CID 57558522.
- ^ Pawlak A (2005). "Inspirierende Proteine". Physik Journal. 4: 12.
- ^ "Julian Voss-Andreae Sculpture". Retrieved 2007-06-14.
Further reading
[edit]- Pieribone V, Gruber D (2006). Aglow in the Dark: The Revolutionary Science of Biofluorescence. Cambridge: Belknap Press. ISBN 978-0-674-01921-8. OCLC 60321612. Popular science book describing history and discovery of GFP
- Zimmer M (2005). Glowing Genes: A Revolution In Biotechnology. Buffalo, NY: Prometheus Books. ISBN 978-1-59102-253-4. OCLC 56614624.
External links
[edit]- A comprehensive article on fluorescent proteins at Scholarpedia
- Brief summary of landmark GFP papers
- Interactive Java applet demonstrating the chemistry behind the formation of the GFP chromophore
- Video of 2008 Nobel Prize lecture of Roger Tsien on fluorescent proteins
- Excitation and emission spectra for various fluorescent proteins
- Green Fluorescent Protein Chem Soc Rev themed issue dedicated to the 2008 Nobel Prize winners in Chemistry, Professors Osamu Shimomura, Martin Chalfie and Roger Y. Tsien
- Molecule of the Month, June 2003: an illustrated overview of GFP by David Goodsell.
- Molecule of the Month, June 2014: an illustrated overview of GFP-like variants by David Goodsell.
- Green Fluorescent Protein on FPbase, a fluorescent protein database
- Overview of all the structural information available in the PDB for UniProt: P42212 (Green fluorescent protein) at the PDBe-KB.
