Coronavirus 3′ stem-loop II-like motif (s2m)
 From Wikipedia the free encyclopedia
From Wikipedia the free encyclopedia
| Coronavirus 3′ stem-loop II-like motif (s2m) | |
|---|---|
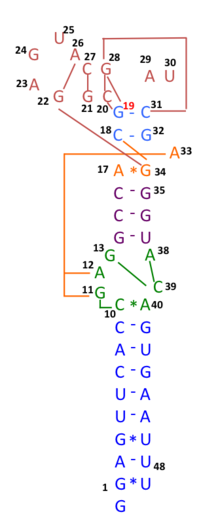
 Predicted secondary structure and sequence conservation of s2m | |
| Identifiers | |
| Symbol | s2m |
| Rfam | RF00164 |
| Other data | |
| RNA type | Cis-reg |
| Domain(s) | Eukaryota; Viruses |
| SO | SO:0000233 |
| PDB structures | PDBe |
The Coronavirus 3′ stem-loop II-like motif (also known as s2m) is a secondary structure motif identified in the 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes.[1][2] Its function is unknown, but various viral 3′ UTR regions have been found to play roles in viral replication and packaging.
This motif appears to be conserved in both nucleotide sequence and secondary structure folding indicating a strong evolutionary selection for its conservation. The presence of this conserved motif in different viral families is suggested to be the result of at least two separate recombination events.[2] To date s2m has been described in four families of positive sense single-stranded RNA viruses; Astroviridae, Caliciviridae, Picornaviridae and Coronaviridae. The viruses that contain s2m can infect a wide range of higher vertebrates, including birds, bats, horses, dogs and humans, and display different tissue tropisms.[3][4] There seems to be a xenologue of s2m in a number of only distantly related insect species.[5]
Other RNA families identified in the coronavirus include the coronavirus frameshifting stimulation element, the coronavirus packaging signal and the coronavirus 3′ UTR pseudoknot.

Biological significance
[edit]Functionally during host invasion by viral RNA, it appears that s2m first binds one or more proteins as a mechanism for the viral RNA to substitute host protein synthesis. This has also been seen in s2m RNA macromolecular substitution of ribosomal RNA folds. The s2m RNA element are also effective targets for the design of anti-viral drugs and antisense oligonucleotides.[6][2]
Potential interacting human microRNA targets of SARS-CoV-2 that share similarity with those of influenza A virus H1N1 was identified as therapeutic targets.[7]
Different structure between SARS-CoV-1 and SARS-CoV-2 s2m
[edit]The overall X-ray (2.7-Å) crystal structure of the s2m SARS-CoV-1 RNA and [2] is different from the SARS-CoV-2 s2m secondary structure determined by NMR.[8] Long-distance interactions between the 5′ UTR and s2m in SARS-CoV-2 genomic RNA have been suggested.[9]
Mutations of SARS-CoV-2 s2m: recombination hotspots rather than a conserved RNA motif
[edit]During COVID-19 pandemic in 2020, many genomic sequences of Australian SARS‐CoV‐2 isolates have deletions or mutations (29742G>A or 29742G>U; "G19A" or "G19U") in the s2m, suggesting that RNA recombination may have occurred in this RNA element. 29742G("G19"), 29744G("G21"), and 29751G("G28") were predicted as recombination hotspots.[10] 29742G>U mutation was also linked to travellers returning from Iran to Australia and New Zealand.[11] In three patients in Diamond Princess cruise, two mutations, 29736G > T and 29751G > T ("G13" and "G28") were found in s2m of SARS-CoV-2, as "G28" was predicted as recombination hotspots in Australian SARS-CoV-2 mutants. This result suggests that s2m of SARS-CoV-2 is RNA recombination/mutation hotspot rather than a conserved RNA motif found in other coronaviruses.[12]
Molecular dynamics simulations shows that both S2M variants, 29734G>C (G11C) and 29742G>U (G19U), of SARS-CoV-2 change RNA structure stability, raising questions as to the functional relevance of s2m in SARS-CoV-2 replication.[13]
See also
[edit]- Coronavirus 5′ UTR
- Coronavirus 3′ UTR
- Coronavirus 3′ UTR pseudoknot
- Coronavirus frameshifting stimulation element
- Coronavirus packaging signal
References
[edit]- ^ Jonassen CM, Jonassen TO, Grinde B (April 1998). "A common RNA motif in the 3' end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus". The Journal of General Virology. 79 ( Pt 4) (4): 715–8. doi:10.1099/0022-1317-79-4-715. PMID 9568965.
- ^ a b c d e Robertson MP, Igel H, Baertsch R, Haussler D, Ares M, Scott WG (January 2005). "The structure of a rigorously conserved RNA element within the SARS virus genome". PLOS Biology. 3 (1): e5. doi:10.1371/journal.pbio.0030005. PMC 539059. PMID 15630477.
- ^ Tengs T, Kristoffersen AB, Bachvaroff TR, Jonassen CM (April 2013). "A mobile genetic element with unknown function found in distantly related viruses". Virology Journal. 10 (1): 132. doi:10.1186/1743-422X-10-132. PMC 3653767. PMID 23618040.
- ^ Rangan R, Zheludev IN, Hagey RJ, et al. (August 2020). "RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look". RNA. 26 (8): 937–959. doi:10.1261/rna.076141.120. PMC 7373990. PMID 32398273.
- ^ Tengs T, Delwiche CF, Monceyron Jonassen C (March 2021). "A genetic element in the SARS-CoV-2 genome is shared with multiple insect species". The Journal of General Virology. 102 (3). doi:10.1099/jgv.0.001551. PMC 8515862. PMID 33427605.
- ^ Lulla V, Wandel MP, Bandyra KJ, et al. (June 2021). "Targeting the Conserved Stem Loop 2 Motif in the SARS-CoV-2 Genome". J Virol. 95 (14): e0066321. doi:10.1128/JVI.00663-21. PMC 8223950. PMID 33963053.
- ^ Chan AP, Choi Y, Schork NJ (November 2020). "Conserved genomic terminals OF SARS-COV-2 as co-evolving functional elements and potential therapeutic targets". mSphere. 5 (6): e00754-20. doi:10.1128/mSphere.00754-20. PMC 7690956. PMID 33239366.
- ^ Rangan R, Watkins AM, Chacon J, et al. (March 2021). "De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures". Nucleic Acids Res. 49 (6): 3092–3108. doi:10.1093/nar/gkab119. PMC 8034642. PMID 33693814.
- ^ Ziv O, Price J, Shalamova L, et al. (December 2020). "The Short- and Long-Range RNA-RNA Interactome of SARS-CoV-2". Molecular Cell. 80 (6): 1067–1077. doi:10.1016/j.molcel.2020.11.004. PMC 7643667. PMID 33259809.
- ^ Yeh TY, Contreras GP (July 2020). "Emerging viral mutants in Australia suggest RNA recombination event in the SARS-CoV-2 genome". The Medical Journal of Australia. 213 (1): 44–44.e1. doi:10.5694/mja2.50657. PMC 7300921. PMID 32506536.
- ^ Eden JS, Rockett R, Carter I, et al. (April 2020). "An emergent clade of SARS-CoV-2 linked to returned travellers from Iran". Virus Evolution. 6 (1): veaa027. doi:10.1093/ve/veaa027. PMC 7147362. PMID 32296544.
- ^ Yeh TY, Contreras GP (1 July 2021). "Viral transmission and evolution dynamics of SARS-CoV-2 in shipboard quarantine". Bull. World Health Organ. 99 (7): 486–495. doi:10.2471/BLT.20.255752. PMC 8243027. PMID 34248221.
- ^ Ryder SP, Morgan BR, Coskun P, et al. (May 2021). "Analysis of Emerging Variants in Structured Regions of the SARS-CoV-2 Genome". Evol Bioinform Online. 17: 11769343211014167. doi:10.1177/11769343211014167. PMC 8114311. PMID 34017166.
